An Update on the Methodology Hepatitis B Virus (HBV) Uses for its Lifecycle: from Nuclear Import of Capsid to ccc DNA for Permanent HBV Cure Acquisition /Avoidance of Persistence of ccc DNA
Kulvinder Kochar Kaur1, Gautam Nand Allahbadia2 and Mandeep Singh3
1Scientific Director, Centre For Human Reproduction, Scientific Director cum Owner Dr Kulvinder Kaur Centre For Human Reproduction, 721,G.T.B. Nagar,Jalandhar-144001,Punjab,India
2Scientific Director, Ex-Rotunda-A Centre for Human Reproduction 672, Kalpak Garden,Perry Cross Road, Near Otter’s Club, Bandra(W)-400040 MUMBAI,INDIA
3Consultant Neurologist Swami Satyanand Hospital Near Nawi Kachehri, Baradri, Ladowali road, JALANDHAR, PUNJAB.
*Corresponding author: Kulvinder Kochar Kour, Centre For Human Reproduction, Scientific Director cum Owner Dr Kulvinder Kaur Centre For Human Reproduction, 721, G.T.B. Nagar, Jalandhar-144001, Punjab,India
Citation: Kour KK, Allahbadia GN, Singh M. (2023) An update on the Methodology Hepatitis B Virus (HBV) Uses for its Lifecycle: From Nuclear Import of Capsid to ccc DNA for Permanent HBV Cure Acquisition /Avoidance of Persistence of ccc DNA. Genesis J Microbiol Immunol.1(1):1-21.
Received: July 11, 2023 | Published: August 6, 2023
Copyright© 2023 by Kour KK, et al. All rights reserved. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Hepatitis B virus (HBV) continues to be a main global public health issue with a determined >250million chronically infected individuals at a risk of generation of chronic liver diseases inclusive of cirrhosis & Hepatocellular carcinoma (HCC). Previously we had reviewed the part of CRISPR/Cas9 system in HBV treatment besides role of epigenetic treatments for DKD; generating /epigenetic control of ccc DNA, in detail for silencing ccc DNA forever. Never the less it is dissapointing that despite efforts, viral elimination does not take place. Despite, antiviral therapies displayed effectiveness in the controlling of virus replication, & enhance liver function, they are incapable of curing HBV infection.
HBV possesses a partly double stranded relaxed circular DNA (rc DNA) genome which can be transformed into covalently closed circular DNA (to ccc DNA) in the nucleus of the infected hepatocytes by cellular DNA healing machinery. ccc DNA correlates with nucleosome to generate-beautified minichromosome which transcribes RNA to validate the expression of viral proteins & reverse transcriptional replication of viral DNA. Apart from the’de novo’’ generation from the incoming virionrc DNA, ccc DNA generation can further take place in the progeny of nucleocapsids with in the cytoplasmic of infected hepatocytes through the intracellular amplification pathway. For these reasons that is sustenance of virus circular episomal lDNA alias ccc DNA in the nuclei of the infected cells & continuation. Apart from hoodwinking all antiviral therapies, ccc DNA further finds a way of evading innate immunity. This viral intermediate possesses a central part in HBV replication, in view of ccc DNA working in the form of a template for transcription of total viral RNA’s inclusive of pregenomic RNA (pgRNA) which in turn feeds the generation of ccc DNA via a step of reverse transcription. Hence the generation /or expression of the ccc DNA is the basic target for eradicating HBV. We detailed newer inventions over role of precure protein, HSP in continuation etc.
Keywords
Hepatitis B virus (HBV); Covalently closed circular DNA (to ccc DNA); nuclear import; Nuclear pore complex (NPC)
Introduction
The human Hepatitis B virus (HBV) continues to be a big hurdle with a determined 250-291million individuals that are chronically infected all over the world [1]. According to epidemiological studies it has been determined that continuation of HBV infection constitutes the main risk factor for the generation of Hepato cellular carcinoma(HCC), pointing to HBV as 1of the maximum significant environmental carcinogens regarding humans [2]. HBV is the archetype members belonging to the family of small enveloped DNA viruses known as hepadna viruses that has a predeliction for infecting hepatocytes. The whole family of hepadnaviruses possess the capacity of replication of their genome in the cytoplasm via reverse transcription of the encapsulated pregenomic RNA (pg RNA) via the viral reverse transcriptase (Pol) (Figure1) [3].
Figure 1: Courtesy ref no-3-Schematic representation of the human hepatitis B virus (HBV) life cycle. After binding to the high affinity receptor sodium taurocholate co-transporting polypeptide (NTCP), HBV is internalized via endocytosis, and the viral capsid is released into the cytoplasm. The capsid travels to the nuclear pore using the microtubule network. The rcDNA is released in the nucleus and is converted into covalently closed circular DNA (cccDNA), which becomes chromatinized, and is the template for the transcription of all HBV RNAs. In the cytoplasm, pgRNA is encapsidated and retrotranscribed into rcDNA. The capsids are then either enveloped at MVB and secreted as new virions, or transported back to the nuclear pore to increase the pool of cccDNA. The steps of the viral life cycle discussed in the review are delineated in the red box.
This enzyme possesses DNA-based DNA polymerase in addition to RNA based DNA polymerase actions along with RNaseH activity which produces a partly double stranded DNA genome known as the relaxed circular DNA (rc-DNA)[4].The 32kbrc-DNA gets encircled in an icosahedral nucleocapsid which is enveloped to generate an infectious particles. On infection, the healing of rc-DNA intermediate kind takes place to form a total double stranded covalently closed circular DNA (ccc DNA)whose placement is in the nucleus which works in the form of a template for the transcription of the viral RNA ‘s inclusive of pg RNA[5].In the cytoplasm; where subsequent to reverse transcription, assembly of mature capsids possessing the rc-DNA takes place with the viral enveloped at the cellular multivesicular body, aiding in the generation as well as liberation of viral particles (see Figure1)[6-7].
In an alternate fashion, capsids might reach back directly to the nucleus leading to ccc DNA getting augmented [8]. There still is dearth of clarification of mode which guides the viral capsids to any of these pathways. Intracellular recycling apparently subsequent to early infection, prior to virus liberation for the associated duck Hepatitis B virus [9]. Thereby it has been pointed that the capsids ultimate fate gets regulated by viral envelope proteins [10]. The development of ccc DNA in the nucleus needs the finishing of various steps i) initiated with the anchoring of the viral particles at cell surface ii) with subsequent transportation of the nucleocapsids to the cytoplasmiii) translocation of the rc-DNA into the nucleus, in addition to its healing for the generation of ccc DNA. The precise narration in the clarity of latter 2 step sis not there as well as might overlap as described further. Noticeably, apart from liberation of viral particles which possess the rc-DNA possessing virions, separate kinds of incomplete capsids which are observed in case of infected humans 100 times or greater [11]. These kinds of empty viral particles has invoked greater investigations by separate groups in view of them portraying a plausible biomarkers with regards to monitoring intrahepatic HBV reactions to antiviral treatments[12]. HBV RNA possessing capsids are further liberated in the form of both non enveloped in addition to enveloped particles [13].
The present therapies for Chronic Hepatitis B virus regulate virus replication along with result in improvement of liver function with effectiveness; however they do not result in total viral clearance in addition to need for lifelong treatment for such patients[14]. The major reasons for failure of treatment is partly the incapacity of the drugs to deplete the ccc DNA completely, which makes it a crucial target for attaining HBV cure. Attaining insight with regards to HBV ccc DNA biology right from viral particles gaining entry to the development of ccc DNA in addition to transcriptional control along with continuation varies & might aid in the isolation of new therapeutic targets for clearance of viral reserve, there by portraying a substantially fertile field of research .Earlier we had reviewed the part of CRISPR/Cas9 system regarding treatment of HBV as well as role in treatment of other diseases, besides role of epigenetic treatments for Diabetic Kidney Disease(DKD), along with in placental dysfunction [15-19] along with reviewed the generation besides the epigenetic controlling of the ccc DNA macrochromosome, the manner host as well as viral factors impact transcription besides if utilization of epigenome editing could be done for silencing HBV ccc DNA forever. Here our aim was to summarize the early processes of HBV infection, with concentration regarding the key steps from the nuclear import of the nucleo capsids to ccc DNA generation with the idea of having insight in ccc DNA generation to avoid its continuation.
Methods
Here we conducted a systematic review utilizingsearch engine pubmed,google scholar ;web of science ; embase; Cochrane review library utilizing the MeSH terms like CHB; HBx; HBc; ccc DNA minichromosome; intracellular trafficking ; intracellular ccc DNA amplification ; viral factors; host factors; viral replication; Hepatocellular carcinoma(HCC) ; Epigenetics; DNA methylation; Histone post-translational modifications; DNA methylation; Histone acetylation ; Histonedeacetylasel; zinc finger nucleases; (TALEN); (CRISPR) CRISPR /Cas9; Designer nuclease(s) from 1985 till date in 2023july.
Results
We found a total of 500 articles out of which we selected 93 articles for this review. No meta-analysis was done.
Entry of virus
The viral envelope possesses 3 viral surface proteins: namely) the large Hepatitis B surface protein Lii) the middle Hepatitis B viral protein,M iii) the small Hepatitis B viral surface proteinS. Initially HBV anchors with a lesser affinity to hepatocytes through binding to heparan sulfate Proteoglycans (HSPG’s), that gets followed by binding to the greater affinity receptor Na+-taurocholate co-transporting polypeptide (NTCP). The evaluation of HBV internalization in addition to the modes controlling it were considerably restricted for numerous decades in view of the isolation of NTCP in the form of HBV receptor revelation as late as just 9 yrs. back [20]. Whereas HBV gains entry to the cell through endocytosis; it is stll debatable, what is the precise mode -if caveola or clathrin based [21]. Studies performed recently confirmed the latter as the main mode implicated in HBV entry [22]. Furthermore, it has been illustrated that the machinery implicated in the endocytosis of epidermal growth factor receptor (EGFR), that works in the form of cofactor for HBV entry is further implicated in the internalization along with transport of the endosomal network of HBV [23]. The modes resulting in HBV escape from endosomes, still need clarification as well as membrane fusion might be modulated via the fusogenic domains isolated in the separate surface proteins L,M as well as S [6-24].
Intracellular Transport along with the Nuclear Import
On liberation in the cytoplasm, the HBV nucleocapsid would be directed towards the nucleus to liberate the viral genome. The HBV capsid gets constituted mainly of 120 dimers of the HBV core protein (HBc) that might assort into a T=4icosahedral symmetry [7]. Based on the virus genotype HBc is comprised of 183/185 amino acids in addition to possesses 2 different domains, N-terminal domain (NTD) implicated in the generation of the capsid shell as well as the C-terminal domain (CTD) which has enrichment of arginine residues as well as are imperative for crosstalk with nucleic acids. CTD is responsible for pg RNA encapsidation as well as DNA maturation at the time of HBV replication [25]. Nevertheless, there is oversimplification of the concept of functional segregation of HBc, in view of various studies have illustrated that NTD possesses a part in DNA generation, whereas on the other hand, CTD takes a part in assembling capsids [26]. CTD is remarkably basic in addition to goes through dynamic phosphorylation along with dephosphorylation that is responsible for control of numerous HBc functions inclusive of pg RNA encapsidation, reverse transcription, stability of capsids along with intracellular trafficking (alias occurs between subcellular compartments like Golgi cisternae and multivesicular endosomes for transport of soluble proteins as MVs. Budding of MVs directly from plasma membrane as microvesicles released outside the secretory cells) [27].
Since non proliferating hepatocytes get infected by HBV, for the HBV genome to gain entry into the nucleus it needs transition via nuclear pore complex (NPC), that works in the form of a watchman for the nucleus. NPC is constituted of numerous copies of huge quantities of proteins known as nucleosporins (Nups), producing a structure which spans the nuclear envelope which comprises of dual membrane. NPC generates channels which aids passive diffusion of small proteins(<40kDa), whereas excluding the bigger proteins. The nuclear import of these bigger protein simply indicates the crosstalk amongst nuclear import receptor members of the import in proteins Imp/karyopherin family. The crosstalk gets stimulated by the recall of a nuclear targeting signal on the surface of cargo proteins by the import receptor [28]. Akin to other viruses, HBV takes that advantage of the microtubular networks for effective nuclear transport for getting over the greater cytoplasmic viscosity. Furthermore, microtubules-based motion might give provision of a direct as well as effective route towards the NPC in addition to the pool of import in proteins having placement at the nuclear periphery [29]. Studies have illustrated that the destabilization of microtubules with the utilization of nocodazole leads to blockade of the HBV capsids in addition to correlated duck Hepatitis B virus (DHBV)[30]. The Group of Kann M, further gave evidence of the need of a microtubule for HBV nuclear trafficking as well as demonstrated that the HBV utilizes the dynein motor complex for nuclear trafficking through a crosstalk with the dynein light chain LL1 [31].
The mode resulting in rc-DNA nuclear import is a matter of controversy, with2 separate models having been posited. According to the1st model, HBV capsids are thought to be implicated in NPC docking as well as genome transport. Actually, the CTD area is considerably preserved amongst all genotypes, possess nuclear localization signal (NLS) sequences which have been revealed to be functional on utilization of NLS abnormal SV 40 large T antigenin the form of a reporter for subcellular placement or by evaluating the influence of mutationon HBc subcellular placement [27,32]. The part of such sequences in nuclear trafficking further supports with regards to assembling of capsidsby conducting competition experiments regarding NPC binding with peptides correlating with NLS utilizing digiton in – permeabilized cells or by asssessment of subcellular placement in hepatoma cells that got cotransfected with a vector coding HB cpossessing mutations in the acknowledged NLS in addition to viral polymerase [3-33]. Furthermore, studies with the utilization of co immune precipitation assays or nuclear binding assessment along with competition assays with wheat germ agglutinin (WGA), that results in active nuclear import blockade by nuclear transport receptors importinα (Impα) as well as importinβ(Imp β) [33-5]. Noticeably, the precise location of the NLS sequences is a matter of controversy, however apparently it overlaps with the 2 of the arginine enrichment domains at residues150-152 as well as 165-168[33-35]. These findings suggest a conformation, where the HBc CTD possessing the NLS should get exposure to the external part of the capsid. The exposure of CTD might differ based on the state of HBc phosphorylation, the kind of nucleic acids existing along with the step of genome maturation [35]. Actually, biochemical along with cryoelectron microscopy (cryoEM) Structural studies have illustrated that based on if RNA is there or not (empty or pg RNA filled capsids)in addition to the status of phosphorylation) with utilization of phosphomimetic CTD mutated), CTD expression is differential [26-32-33].Thereby CTD’s exposure in the exterior to an empty capsid is transitory; whereas the negatively charged RNA keeps the basic CTD within the capsid[36]. Nevertheless, the phosphorylation of the exposure capsid possessing RNA has been illustrated to stimulate nuclear pore binding, confirmed the posit that phosphorylation modulates CTD exposure [35]. The capsids of mature HBV particles (particles filled with rc-DNA) apparently is basically dephosphorylated; that as per theory could result in CTD getting retained in the interior. Nevertheless, the maturation events via which RNA gets retro transcribed into new ds-DNA is correlated with structural alterations. Hence mature capsids are considerably unstable, partly in view of kind of DNA, that possesses lesser flexibility in addition to possesses considerably lesser capacity of crosstalking with CTD’s[37]. CTD’s thus might get exposure to the external part of the capsid at the time of breathing [38]. Furthermore, as previously described phosphorylation portrays a dynamic event which can further be implicated in CTD exposure in mature capsids. [39], recently detailed the phosphorylation of a mature capsid that takes place on viral entry or recycling of capsids in addition to that might further modulate trafficking, uncoating as well as ccc DNA generation [39]. Lastly CTD’s exposure in mature particles is validated by studies illustrating that mature capsids undergo cleavage by trypsin in contrast to immature particles having part protection from digestion [35]. Whereas pg RNA possessing capsids have been illustrated to basically get exclusion from NPC; probably in view of CTD’s placement in the interior along withthereby do not have accessibility for importin crosstalk; cryoEM studies have illustrated that CTD’s in the empty capsids get exposure at the external aspect of the capsids [35-29]. Nevertheless, in the empty capsids CTD’s crosstalk directly with importin β(Imp β) through an importin β binding sequence(IBB) with placement amongst amino acids 141-180,implying a unique intracellular traffickingin them in contrast to mature capsids [40]. If Imp β is implicated in trafficking of empty capsids in addition to capsid destabilization, as pointed in this study needs future evaluation.
Subsequent to their docking to NPC’s, HBV capsids move to the nuclear basket where uncoating takes place through capsid disassembly. This is followed by HBV DNA liberation into the nucleoplasm as well as HBc dimers, that possess the capacity of recorrelating to generate capsids [41]. At the time of their study Kann M’s group found an unexpected characteristics of nuclear import : despite immature as well as mature capsid crosstalk with the NPC along with the diffusion in the nuclear basket, just mature capsids breakdown whereas immature capsids continuously are trapped. They illustrated this trapping was brought about by direct cross talk of the viral capsid NTD with a protein of the nuclear basket; Nucleosporins 153(Nup153). Intriguingly,mature capsidsat the time of binding with Nup153 breakdown, aiding capsid dimers with viral DNA to gain access to nucleoplasm (Figure2)[42].
Figure 2: Courtesy ref no-3-Models of HBV genome nuclear import. (1) Capsid disassembly/partial disassembly occurs in the cytoplasm (or at the nuclear pore) leading to the exposure of the nuclear localization signal (NLS) in the terminal protein (TP)-domain of Pol, and the subsequent binding to nuclear transport factors (importin α and β). (2) Capsids are transported into the nuclear basket following the interaction of NLS present in the core CTD with the import factors importin α/β. There, capsids bind directly to Nup153 and mature capsids disintegrate, leading to the release of the viral genome. Capsid dimers enter the nucleus where they can re-assemble. rcDNA is represented as attached to the viral polymerase (red circle). Capsid dimers are represented as purple triangles.
The direct binding of the capsid to Nup153 might offer an explanation with regards to Ran GTP is not needed for the liberation of the viral DNA despite the capsid having been bound to the importin α/β at the time of the initial step of translocation. The query arises -the precise advantage of binding with Nup153once capsids gain access through importin α/β transportation as well as if there is need of transitory binding of capsids with Nup153 regarding their breakdown. Apparently, disassembly takes place in the nuclear basket since UV cross linking of capsids does not result in dysfunctional placement of capsids in the basket, where as it results in dysfunction of the HBV DNA nuclear translocation. [42]. The modes resulting in uncoating requires clarification. The way described previously mature capsids possess lesser stability. In vitro experiments have illustrated that capsids filled with ds DNA possess lesser stability in contrast to capsids correlated with RNA [7-37]. DNA generation might aid in capsid destabilization in view of DNA’s properties, that have greater rigidity. Post-translational modifications of the capsid like phosphorylation might further promote uncoating [26-37]. Actually, sustenance of capsid stability is thought to be via the formation of balanced electrostatic crosstalk in the capsid that might be modulated by phosphorylation/de phosphorylation processes [43]. Furthermore, host cellular factors might further be implicated in the selective events of mature capsids uncoating. Actually, mouse cells do not validate HBV ccc DNA generation. Nevertheless, one mouse hepatoma cell line has recently been illustrated to result in accrual of HBV ccc DNA that associates with the unstable capsids in the cytoplasm [44].Taking into account that there is incapacity of mouse cells to aid in ccc DNA generation is not secondary to the expression of murine restriction factors; however more probably in view of the lack of cellular factors one can posit that HBV uncoating, further needs the actions of host factors whose identification is still awaited [45].
Moreover, the opening of the capsid in the nuclear basket might further be advantageous for the virus. Actually, it has been illustrated that the nuclear basket acts in the form of a platform for DNA healing in addition to possesses enrichment of cellular factors implicated in DNA healing [46]. Thereby the liberation of rc DNA in the nuclear basket might facilitate its healing in addition to aid the virus to escape the cellular reactions which might be stimulated in case if rc DNA got directly liberated to nucleoplasm. There is need for future studies to find if nuclear pore basket might be significant with regards to ccc DNA generation escaping along with for antiviral cellular reaction escape. Nevertheless, [47], hypothesized that viral import gets modulated by the viral polymerase which possesses a bipartite nuclear localization signal (NLS)[63]. Import modulated by the polymerase would need the part/full exposure of the polymerase the cytoplasm or at the cytoplasmic face of the NPC(Figure2). The existence of an unstable or part/ disassembled capsid) in the cytoplasm continues to be controversial. Guo JT ‘sgroup has found the existence of DNase -1 sensitive nucleocapsid (pointing to a greater unstable or part/ disassembled capsid) in the cytoplasm. Maximum of these capsids as per their outcomes possess deproteinated rc DNA (dprc DNA) which they believe are precursors of ccc DNA [31-45-46]. They pointed that the maturation events with in the capsid (finishing of the (+) strand DNA along with elimination of the polymerase) in the cytoplasm results in structural alteration, aiding in the exposure of the capsid NLS as well as nuclear import [46]. Nevertheless, their outcome does not reveal with clarity if the dprc DNA is the actual precursors of ccc DNA or a dead-end product. Furthermore, their observations were queried by other researchers whose findings were that cytoplasmic capsid sin general possess resistance to DN ase digestion [48]. Future studies would be required to detect where the maturation steps resulting in formation of functional precursors of ccc DNA take place. Nevertheless, evaluating the Guo JT ‘s group outcomes one cannot completely discount the presence in the cytoplasm of destabilized/ disassembled capsids possessing polymerase bound rc DNA [49]. Actually, in coimmunoprecipitation experiments with the utilization of antibodies against capsid C terminal amino acid residues or karyopherin, dprc DNA recovery portrays just a percentage of the total precipitated DNase -1 sensitive DNA pointing that partial of this is still viral polymerase bound. Here PolNLS might further be implicated in the nuclear import. Full disassembly of the capsid as well as the liberation of the viral DNA bound to polymerase in the cytoplasm apparently is not likely; in view of the viral DNA getting sensed by the innate immune system [50].
Transformation of rc DNA to ccc DNA
rc DNA possesses a unique structure correlated with the manner it is generated by the viral polymerase (Pol). 1stly as compared to the canonical reverse transcription, a tyrosine residue (Tyr68) in the terminal protein (TP ) domain of Pol works in the form of acceptor for the 1stdNTP.This kind of ‘’protein priming’’ results in covalent linking of Pol to the 5’ end of the (-) strand of rc DNA. The priming of (+) strand takes place by an 18 nucleotide long capped oligomer obtained from the5’ end of thepg RNA that continues to be correlated with the 5’ end of the (+) strand in the mature DNA. The (+) strand DNA formation closes the gap amongst5’ along with 3’ end of the (-) strand of DNA forming a short triple stranded area known as ‘’r’’. Lastly the (+) strand DNA formation remains incomplete, keeping around 50% of the genome single stranded before the virion production. Noticeably, the total genome maturation steps subsequent to priming need the encapsulation of Pol as well as the viral genome into the capsid with. Summarizing this rc DNA comprises of a complete coding (-) strand covalently anchored at its 5’ end to the viral polymerase as well as bearing a nine nucleotide unnecessary sequences at its extreme ends in addition to harmonious incomplete (+) strand which is conjugated at the5’ end to a capped RNA primer(fig3) [4-51].
Figure 3: Courtesy ref no-3-Schematic representation of the conversion of rcDNA into cccDNA. cccDNA formation requires: (i) the removal of Pol from the 5′ of the (−) strand DNA, (ii) the completion of the (+) strand, (iii) the removal of the capped RNA from the 5′ end of the (+) strand DNA, (iv) the cleavage of the redundant sequence “r”, and (v) the ligation of the 5′ and 3′ ends of the two DNA strands. cccDNA is then loaded with histones. The precise time sequence of these different steps is not yet completely determined.
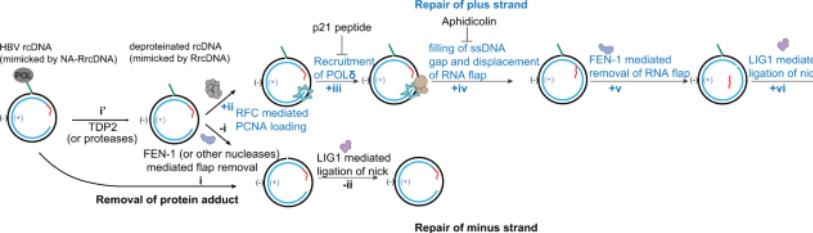
The requirement of ccc DNA production from rc DNA are i)finishing the single stranded part of the viral genomic) the elimination of Pol fromthe5’ end of the (-) strand of DNA iii) the elimination of capped RNA fromthe5’ end of the (+)strand of DNA as well as iv) one strand off’s’’-the dispensable nine nucleotide long triple stranded area, needs to be depleted for aiding in the ligation of 5’ along with 3’ end of the 2 DNAs trends (figure3).These total steps need various enzymes, like DNA polymerases-an endonuclease for elimination of the viral polymerase as well as RNA primer in addition to the DNA ligase. Continuation of being non evaluated for>20-30yrs in view of the absence of a cellular model of infection which was easy to control, the modes implicated in the ccc DNA production we are only recently getting greater insight; however still it needs greater assessment. To begin with the mode implicated in started Pol depletion continues to be controversial. Initiation with the utilization of biochemical in addition to silencing strategies ,NassalM’s group revealed that the cell ulartyrosy lDNA phosphodiesterase 2(TDP2)is the factor implicated in the depletion of the viral polymerase [52].Nevertheless, various groups with the utilization of silencing strategies could not replicate these outcomes[43,67].Recently ,a study having the objective of evaluating the properties of the 4 termini of the cytoplasmic dprc DNA illustrated that the elimination of the viral polymerase takes place via unlinking the tyrosyl DNA phosphodiesterase bond amongst the viral polymerase as well as (-) strand DNA by enzymes which still are required to be unraveled [53].Nevertheless, as stated earlier no clarity exists in the context of if cytoplasmic dprc DNA previously portray the actual intermediate of nuclear ccc DNA. Akin to that, finishing of the (+) strand has been controversial. Whereas studies have illustrated that the viral polymerase is involved in (+) strand finishing, other studies with the utilization of particular hampering agents have made this posit null &void [54-55]. Rather than that (+) strand HBV DNA is finished via the DNA polymerase kappa (Polκ) [55]. Weil as well as Ploss A recently generated an in vitro system with the utilization of recombinant rc DNA as well as cellular extracts from yeast or hepatoma cells for evaluating ccc DNA generation. Identification of cellular proteins responsible for DNA lagging strand production along with DNA healing -the i) proliferating cell nuclear antigen (PCNA), ii) replication factors C complex (RFC) in addition to, iii) DNA polymerase-δ(Pol δ)[56]. Whereas 2the studies isolated 2 separate polymerases machinery, they are not essentially controversial in view of both the polymerases possess the capacity of functioning together at the time of nucleotide-excision repair (NER) [57]. Intriguingly Polκis thought to be significant for DNA healing once the quantities of pool of dNTP are lesser, like in cells that lie in quiescence [58]. Recently, a study by Tang etal.[59], emphasized that numerous modes might be implicated in ccc DNA generation; with them positing that in contrast to production of ccc DNA subsequent to de novo infection, the ccc DNA generation needs the actions of DNA polymerase alpha (Pol α) in addition to Pol δ[59].These interesting results point that rc DNA might vary amongst the rc DNA emanating from de novo infection, in addition torc DNA generated directly from recycling. In an alternate fashion without knowledge why capsids might utilize a separate pathway regarding nuclear importation; thereby the rc DNA might be liberated in separate areas of the nucleus where enrichment of separate DNA healing proteins exists. The in vitro ccc DNA production assay further validated the implication of a factor that had not earlier been isolated by other researchers; namely Flap structure particular endonuclease1(FEN1). FEN1 has been illustrated to be implicated in the cleavage of the unnecessary area ‘’r’’ by the utilization of silencing strategies or a FEN1 hampering agent [60]. Intriguingly Kitamura Tal.[60], with the utilization of an in vitro ccc DNA production assay revealed that transformation of rc DNA into ccc DNA can take place with effectiveness by utilization of Best DNA polymerase DNA ligase1 in addition to FEN1 [60]. Lastly, this in vitro assay further validated the part of LIG1. Actually, with the utilization of RNA screen that covered 107cellular healing genes Long et al. [54], earlier isolated the host cellular ligases1 along with 3 (LIG1 along with LIG3) being the cellular enzymes implicated in rc DNA termini ligation [54]. Furthermore, their screening further isolated 2 DNA healing proteins APEX1 as well as PolB, that might be implicated in ccc DNA production. Additionally, with the utilization of screen dependent on utilizinx`x`g the drugs that target cellular enzymes [61] isolated the two Po isomerases (topoisomerase1 as well as topoisomerase2[TOP1 as well as TOP2] being implicated in the dissemination of viral DNA. Nevertheless, no clarification regarding the mode exists along with for experiments utilizing the si RNA targeting TOP1 controversial outcomes [61]. Whereas, the precise kinetics of ccc DNA production is still not clearly known outcomes from separate laboratories imply that finishing of (-) strand is more rapid in contrast to finishing of (+) strand, apart from occurring independently [62-63].
Another significant query is with regards to the mode which marks the subsequent enrolment of cellular enzymes implicated in ccc DNA production. in view of the injury of DNA like structure of rc DNA, Luo [64], evaluated the role of 2 DNA damage response (DDR) pathways; Ataxia telangiectasia mutated (ATM), as well as Ataxia telangiectasia and Rad3-related (ATR), in ccc DNA production utilizing the hampering agents in addition to silencing strategies. Their outcomes gave evidence regarding the part of ATR in HBV ccc DNA production [64]. In view of the outcomes produced basically in hepatoma cells; it can be queried that the expression of healing proteins as well as pathways actions in cells that lie in quiescence like hepatocytes might vary from cells that are replicating as was illustrated in neurons along with myoblasts [65]. Nevertheless, the liver might not fully belong in this category in view of hepatocytes possess the capacity of sustenance of replication. In view of liver due to its biological functions like detoxification; gets exposure to numerous DNA injuring agents, that need it to be particularly efficient in DNA healing [66]. Nevertheless, evaluating the ccc DNA production in converted cells might possess its biases as well as restrictions. Actually, if conversion is correlated with the breakdown of the DDR pathway; so that their survival is feasible, the cells in general generate modes to escalate their healing ability through another unessential healing pathway [67]. The organization of ccc DNA is in the form of minichromosomal correlated with histone in addition to non histone proteins; however, the precise timing in addition to factors implicated in ccc DNA chromatinization needs full insight. Previous studies have isolated HBV nucleon protein complexes from the hepatoma cell lines HepG2.2.15 with by 4 core histone proteins (H2A, H2B, H3, as well as H4) along with linker histone H1 in addition to HBc correlated with ccc DNA [68]. In vitro experiments with Exonucleases oocyte extracts, reconstituted the chromatin on HBV ccc DNA in addition to rc DNA, despite just the ccc DNA possessed the controlled spacing along with structure [86]. Intriguingly.[68], observed that the binding of the HBc to the ccc DNA is correlated with a diminished spacing of the nucleosomes [68]. Studies performed more recently utilizing DHBV as well as HBV observed that the assembly of nucleosomes into the ccc DNA does not possess random placement as well as are nucleosomes eliminated are as were observed in the Enhancer I as well as Enhancer II/BC Parees [69]. Clarification is still required regarding chromatinization takes place subsequent to nuclear import inrc DNA, or if it occurs only subsequent to healing in addition to transformation of rc DNA to ccc DNA.
Post-translational modifications of histones that have assembled into the ccc DNA takes place along with they control HBV transcription as well as replication, apart from that HBx is significant for the generation as well as the sustenance of an active chromatin status in the ccc DNA [70-72].An assessment of the ccc DNA chromatin displayed an abundance for active histone modifications, specifically the acetylation of H3 and H4 histones apart from trimethylation of histone3 lysine4((H3K4me3), absence of (H3 K27m3) in addition to differing quantities of the repressive marks (H3 K9me3)were observed in samples from chronically Infected HBV patients, pointing to a complicated controlling taking place in chronic HBV infection[54,57]. More recently, Alvarez Astudillo etal.[74], isolated a non-classical histone variantH3.3.2 in the form of a positive controller of HBV ccc DNA [74]. Greater details regarding epigenetic as well as transcriptional control of HBV ccc DNA is available in [ref5,75]. Rarely reverse transcription of the encapsulation does not continue appropriately along with result in the formation of a double stranded linear DNA (dsL-DNA) which can trace the same fate like rc DNA possessing capsids (liberation or intracellular recycling) [4]. In the nucleus, the dsL-DNA HBV genome might be either without efficiency get transformed by non-homologous end joining (NHEJ), into a nonfunctional DNA or get incorporated into the cellular genome by NHEJ or microhomology modulated end joining [54-75]. Nevertheless, the incorporated HBV DNA does not possess replication-competence; however, is thought to be responsible for generation of Hepatocellular carcinoma [76].
Conclusion
HBV has assumed a worldwide problem as well as is implicated in greater than 880,000 mortality rates/year. Present therapy that has received approval for Chronic HBV Infections (CHB) are inclusive of nucleoside /nucleotide analogues, as well as interferon-alpha (IFNα); which enhance liver function. Nevertheless, they do not provide full cure in view of them not implicated in full elimination of ccc DNA resulting in viral continuation. Maximum of our information is obtained from the evaluation of ccc DNA in view of animal or tissue culture models [78]. The quantity of HBV ccc DNA in various in vivo, as well as in vitro models differs however varies among 0.2-2 copies/cell [79]. In a recent well generated HBV infection model aiding in viral progression,5-12 copies were observed in each infected cell, with a half-life determined to be about 40days [10]. Furthermore, HBV ccc DNA was evaluated in CHB patients receiving nucleoside /nucleotide analogues treatment [80]. These studies have illustrated that continuation of ccc DNA takes place in Chronic patients although therapy administration was given with the determined half-life being 9.2 mths. The reasons offered for the sustenance in addition to replenishing of ccc DNA pools might be secondary to continuous replication, that is persistent at the time of treatment [80-81] recently [82], utilized the formation of lamivudine resistant HBV mutants for assessment of ccc DNA turnover. They displayed that ccc DNA turnover was more rapid in contrast to earlier pointing with the variation of half-life from 5.6-21 wks. [82. Thereby [82], implied that the full removal of ccc DNA might be attained with approaches which fully block replenishing of ccc DNA pools. Nevertheless, this study evaluates the turnover of actively transcribing ccc DNA; however, it is not feasible to rule out the presence of practically silent ccc DNA which might continue in the form of pools/reservoirs. Whereas modes of ccc DNA sustenance are still not fully clarified, the illustration fully of the total steps implicated in ccc DNA production from entry of rc DNA to its healing along with expression of ccc DNA, will aid in isolating the therapeutic targets for blockade of newer molecules of ccc DNA. Furthermore, insight regarding modes implicated in capsid translocation of rc DNA liberation as well as the isolation of the cellular factors which crosstalk at the time of early steps might aid in getting insight why sensing of HBV does not take place by innate immune system. Restoring the antiviral reactions might aid in the removal of HBV combined with other anti-viral therapies [83]. Till date various agents are under bio generation which directly target separate stages of HBV life cycle or modulate the innate immune reactions or adaptive immune reactions. One of such agents bulevirtide; a synthetic N- acetyl peptide obtained from HBsAg, blocks HBV entry; having received approval for Chronic HBV Infections (CHB) from European Union. Agents which target nucleocapsids assembly known as capsid assembly modulators (CAM)are in preclinical trials [84]. Canocapavirpor trays one of the novel core protein allosteric modulators (CpAM’s)in phase II trials which stimulates a conformational alteration of the linker area of HBV core protein [85] (Figure 4).
Figure 4: Courtesy ref no-85-The binding site of Canocapavir is located at the HAP pocket. (A) Structural simulation and molecular docking analysis of the binding site of Canocapavir on HBc (PDB: 6J10). (B) HepG2 cells expressing HBc or indicated HBc mutants were mock-treated or treated with 2 μMC enoxaparin for 48 h. Intracellular capsids were analyzed by native gel assay and immunoblotting with anti-HBc C1. The levels of total HBc expression were determined by Western blotting with anti-HBc 2A7. (C) HepG2 cells were transfected with pHBV1.1 or pHBV1.1-derived plasmids with indicated HBc gene mutations, left untreated or treated with 2 µM Bay 41-4109, 2 µM JNJ-6379, or 2 µM Canocapavir, respectively. Intracellular capsids were resolved by native gel electrophoresis and detected with anti-HBc 2A7 or C1, with associated HBV nucleic acids detected by hybridization with a DIG-labeled HBV-specific probe.
Nevertheless, the agreement in this field is that agents would need to be used combined with other anti-viral therapies for attaining HBV cure. Newer therapeutic strategies have been described previously in reviews ref no [83-87], in their attempt to isolate cellular DNA healing protein needed for ccc DNA production with the utilization of chemo genetic screen, observed thatB02 a small molecular hampering agent of DNA homologous recombination (HR) healing protein RAD51, significantly escalated the generation of ccc DNA through the intracellular amplification pathway in human hepatoma cells. Intriguingly, neither small interfering RNA (si RNA) knockdown of RAD51expression nor structurally unique RAD51 hampering agents or activator changed ccc DNA intracellular amplification. Rather it was observed thatB02 treatment significantly escalated the quantities of Heat shock protein (HSP) mRNA as well as si RNA knockdown of RAD51 expression or treatment significantly with structurally unique HSPA hampering agent significantly mitigated B02 escalation of ccc DNA amplification. Furthermore, B02 escalation of ccc DNA amplification was hampered by agents which selectively hamper Pol α or TOPII, an enzyme needed for ccc DNA intracellular amplification was hampered with effectiveness by agents which selectively hamper Pol α or TOPII, thereby their outcomes pointed treatment with B02 stimulates a HSP modulated cellular reaction, which positively control transformation of rc DNA to ccc DNA through the valid intracellular amplification pathway. The significance of this is depletion or functional inactivation of ccc DNA Mini chromosomes in HBV infected hepatocytes is necessary for the cure of Chronic Hepatitis B (HBV) virus infection. Nevertheless, absence of information regarding molecular modes of ccc DNA metabolism, as well as controlling inhibits the formation of anti-viral agents for attaining this therapeutic aim. Their observation displayed here suggest that escalation of ccc DNA amplification might take place in selected pathobiological situations, like cellular stress, to destabilize the dilution or depletion of ccc DNA in addition to sustenance of the continuation of HBV infection. Therapeutic hampering of HSPA escalated ccc DNA amplification in these pathobiological situations needs to promote the depletion of ccc DNA in addition to cure Chronic Hepatitis B [87]. Hepatitis B virus (HBV)is dependent on HBc for generating productive infection in the form of ccc DNA generation in addition to perform each step of the lifecycle subsequent to ccc DNA generation. Numerous HBc copies generate an icosahedral capsid shell which encapsulates the viral pg RNA in addition to promotes the reverse transcription of pg RNA to rc DNA in the capsid. At the time of infection, the full HBV virion that possesses an outer enveloped layer as well as the internal nucleocapsid possessing rc DNA, gains entry in human hepatocytes through an event known as endocytosis along with traffics via an endosomal compartment as well as cytosol for administration of rc DNA into the nucleus to generate ccc DNA. Additionally, progeny rc DNA, newly generated nucleocapsid in cytoplasm is further administered in the same cell for generating ccc DNA in an event known as intracellular ccc DNA amplification or recycling. Recently Mendenhall. [88], illustrated differential actions of HBc in influencing ccc DNA generation at the time of de novo infection vs recycling derived with the utilization of HBc mutations as well as small molecular hampering agents. These outcomes pointed to key part of HBc in estimating HBV trafficking at the time of infection in addition to nucleocapsid disassembly (uncoating)(for liberation of rc DNA ,processes necessary for ccc DNA generation. Probably HBc works in these events through cross talk with host factors which aids crucially to HBV host tropism. Greater insight regarding part of HBc in HBV entry, ccc DNA generation in addition to host species tropism would escalate the continuing work in generating HBV cure as well as promote the formation of animal models which are easy to handle for pursuing basic work in addition to drug generation [88] (Figure5,6).
Figure 5: Courtesy ref no-88-The simplified HBV lifecycle. HBV is endocytosed, traffics through the early (purple) and late (orange) endosomal compartments, undergoes cytosolic release and nuclear import to deliver rcDNA to the nucleus, where it is converted to cccDNA. Transcription and translation then occur, and nucleocapsid packaging of pg RNA and conversion to rcDNA ensues. rcDNA-containing nucleocapsids can be enveloped and secreted or cycled back to the nucleus to make more cccDNA (intracellular amplification). The dashed lines of capsids (hexagons) denote capsid destabilization associated with maturation (rcDNA synthesis), which is thought to prime the mature nucleocapsid for uncoating. The steps impaired (red) or enhanced (green) by HBc mutations and CAMs, or facilitated by species-specific host factors (green), are highlighted. HBV replication steps not directly relevant here are omitted for brevity. Plasmid (HBV replicon) transfection, as depicted, bypasses the entire HBV entry steps to deliver the HBV DNA (cccDNA equivalent) to the nucleus. CAM-A—capsid assembly modulator-aberrant; CAM-E/A—capsid assembly modulator-empty/aberrant; huNTCP—human sodium taurocholate co-transporting polypeptide; ssDNA—single-stranded DNA; rc—rcDNA; ccc—cccDNA.
Figure 6: Courtesy ref no-88-Schematic diagram of HBc domains highlighting residues important for viral entry. HBc residues, in the NTD and linker, shown to play a role in HBV entry are highlighted. Mutations at these can block cccDNA formation during HBV infection but allow/enhance cccDNA intracellular amplification positions. Mutations highlighted in red likely affect cccDNA formation via altered HBc phosphorylation state. Mutations marked in blue affect cccDNA formation via destabilization of nucleocapsids. Mutations listed in black affect cccDNA formation via an unknown mechanism, possibly through cytosolic transport or nuclear import. See text for details.
Furthermore, [ 89], illustrated the HBV precure protein is not necessary for virus replication however has a part in the continuation of virus. In their study they determined numerous precure protein species in HBV infected human hepatocytes as well as evaluated in their function in HBV lifecycle. Their observation was that HBV precure proteins diminished intracellular HBV core protein in addition to decreased liberation of completed virions; however, escalated the liberation of empty virions. Intriguingly, the cytosolic precure protein species generated chimeric capsids with the core proteins along with were liberated in virions. Their results suggested that precure protein species in the HBV lifecycle in addition to had repercussions regarding the part of precore proteins in HBV continuation as well as in pathogenesis [89] (Figure 7).
Figure 7: Courtesy ref no-89-Biosynthesis of HBc and HBV precure protein products. HBV precure and core proteins are expressed from the overlapping precure/core ORFs but using two distinct mRNA templates. HBc, translated from pgRNA, forms the icosahedral capsid inside complete and empty virions. The precure mRNA, which has a 29-aa extension at the 5′ end relative to pg RNA and contains the first initiation codon in the precure/core genes, is the template for translation of the precure protein (p25), the precursor to HBe Ag and Perc The N-terminal signal peptide (SP) of p25 is removed by the signal peptidase translationally during the translocation of p25 into the ER lumen, leading to the production of p22, which is further processed at its CTD before being secreted as the dimeric HBeAg (p17). PreC, which has the noncleaved SP but processed CTD, is also secreted. The exact C-terminal processing sites of HBeAg and PreC are heterogeneous, as indicated by the hashed boxes. The epitopes recognized by the monoclonal antibody (MAb) 1A11, specific to precure-derived proteins (epitope: −10 to 5 aa), MAb T2221 (epitope: 130 to 140 aa), specific to the NTD shared by all precure/core gene products, and MAb 366-2 (epitope: 150 to 164 aa), specific to the CTD of HBc and some precure proteins (p22 and p25), are indicated (blue brackets).
HBV possesses partly double stranded rc DNA genome generated within a nucleocapsid (NC) in the host cell cytoplasm. The liberation of rc DNA from NC, is an ill understood known as uncoating, to the nucleus is needed for its transformation to the ccc DNA with the viral episome working in the form of transcriptional as template for the various transcripts, including pg RNA from which core protein and P protein get translated; the full repertoire of viral RNA essential for replication as well as thereby necessary for production sustenance of infection. For acquisition of greater insight [90], evaluated HBc mutants which illustrated different quantities of nuclear ccc DNA; however little to no cytoplasmic DNA .Their observation was that rc DNA production might take place by these mutants outside however as compared to wild kind(wk.),these mutants NC did not possess the capacity of protecting rc DNA from digestion by the endogenous nucleases in cellular lysates or exogenous DNases .Subcellular fractionation pointed that the main DNA breakdown action was membrane correlated. digestion with sequence particular as well as non-particular DNA ases displayed the exposure of particular areas of rc DNA from the mutant NC. Akin therapy of wk NCs with core hampering agents acknowledged to escalate ccc DNA by influencing uncoating further resulted in area particular exposure of rc DNA. Moreover, a subpopulation of untreated mature NC’s demonstrated area particular exposure of rc DNA also. Competition amongst rc DNA breakdown as well as its transformation to the ccc DNA at the time of NC uncoating; thereby probably possesses a significant part in the formation as well as continuation of HBV Infection in addition to has repercussions on the production of capsid targeted anti-viral. Disassembly of the HBV NC for liberating its genomic DNA; an event-alias uncoating which is not clarified is needed from the viral episomein the host cell nucleus, a viral DNA necessary for formation as well as sustenance of HBV infection. The removal of HBV nuclear episome is the emblem of producing a cure [ 90], revealed that HBV genomic DNA gets exposed in an area particular way at the time of uncoating, that gets escalated by mutations of the capsid targeted protein as well as capsid targeted antiviral agent. The viral genomic exposure can lead to fast breakdown or in an alternate fashion can escalate the generation of the nuclear episome thereby possessing a main influence on HBV infection as well as continuation. These outcomes are significant for getting insight into the foundational modes of HBV replication as well as continuation in the ongoing efforts to find a total cure [90].
HBV pg RNA portrays a circulating biomarker for ccc DNA in HBV infected subjects as well as has been evaluated for the effectiveness of treatment, disease staging along with off treatment results; Nevertheless, there is paucity in the context of stability. Hence enhancing sensitivity of HBV pg RNA assays might escalate its anticipative significance along with give understanding regarding low viral quantities [91], reported how HBV pg RNA v2 assays with escalated sensitivity(152 copies/ml with v1 to 10(0.6ml) along with 22(0.2ml) copies/ml with v2respectively as well as flexible intake volume displayed enhanced determination in addition to quantification of low titer samples .Substantially sensitive HBV pg RNA assays might be of use in refinement of anticipative therapy results dependent on numerous situations, that escalates the dependency of this biomarker[91].
References
- Trepo C, Chan HL, Lok A. (2014) Hepatitis B virus infection. Lancet. 384:2053-63.
- Parkin DM, Bray FI, Devesa SS. (2001) Cancer burden in the year 2000.The global picture. Eur J Cancer. 37(Suppl 8): S4-S66.
- Dias JD, Sarica N, Neuveut C. (2021) Early steps of Hepatitis B life cycle: from capsid nuclear import to ccc DNA formation. Viruses. 13:757.
- Nassal M. (2008) Hepatitis B viruses: reverse transcription a different way. Virus Res. 134(1-2):235-49.
- Turton KL, Meier-Stephenson, Badmalia MD, Coffin CS, Patel TR. (2020) Host transcription factors in Hepatitis B virus synthesis. Viruses. 12(2):160.
- Jiang B, Hildt E. (2020) Intracellular trafficking of HBV particles. Cells. 9(9): 2023.
- Selzer L, Zlotnik A. (2015) Assembly and release of Hepatitis B virus. Cold Spring Harb Perspect Med. 5(12):a021394 .
- Ko C, Chakraborty A, Chou WM, Hasreiter J, Wettengel JM, et al. (2018) Hepatitis B virus genome recycling and de novo secondary infection events maintain stable ccc DNA levels. J Hepatol 69(6):1231-41.
- Tuttleman JS, Pourcel C, Summers J. (1986) Formation of the pool of covalently closed circular viral DNA in hepadnavirus infected cells. Cell. 47(3):451-60.
- Summers J, Smith PM,Horwich AL. (2021) Hepadna virus enveloped proteins regulate covalently closed circular viral DNA amplification. J Virol. 64(6):2819-24.
- Hu J, Liu K. (2017) Complete and incomplete Hepatitis Bvirus particles: formation, function and applications. Viruses. 9(3):56.
- Hong X, Luckenbaugh L, Mendelhall M, Walsh R, Cabuang L, et al. (2021) Hepatitis B precore /corerelated antigen. J Virol . 95(3):e01695-20.
- Shen S, Xie Z, Cai D, Yu X, Zhang H, et al. (2020) Biogenesis and molecular characteristics of serum Hepatitis B virus RNA. PLoS Pathogen . 16(10):e1008945.
- Revill P, Testoni B, Locarnini S, Zoulim F. (2016) Global strategies are required to cure and elimination of HBV infection. Nat Rev Gastroenterol Hepatol. 13(4):239-48 .
- Kaur KK, Allahbadia GN, Singh M. (2021)Potential role of Epigenetic Modulation in prevention or therapy for Diabetic Kidney Disease-still a dream or a reality –A Systematic Review’’.J Diab Nephro Diab Mgmt. 1(1):(1-26).
- Kaur KK, Allahbadia GN, Singh M. (2021) How does Epigenetics Regulate Development of Placenta and Placental Pathologies like PreEclampsia(PE),Intrauterine growth Restriction( IUGR)-With Main emphasis on PE’’.Published in Advances in Bioengineering and Biomedical Science Research.
- Kaur KK, Allahbadia GN,Singh M. (2022) ’An Update on Genome Editing with the Utilization of CRISPR/Cas 9 System for Evaluation and Treatment of Human Diseases-A Systemic Review’’. Curr Trends Biomedical Eng & Bio sci. 20(5).
- Kaur KK, Allahbadia GN, Singh M. (2022) The part played by NonHistone Proteins crotonoylation other than lysine crotonoylation in the Development of Diseases:ASystematic Review’’. Int J Endocrinol Diab. 4(2):124.
- Kaur KK, Allahbadia GN, Singh M. (2022) ’The role of Utilization of Epigenetics Treatment Strategies Regarding Hepatitis B virus covalently closed circular DNA Silencing with the aim of Attaining Cure:A Narrative Review’’ Accepted for Publication in November issue of Acta ASGIS.
- Yan H, Zhang H, Xu G, He W, Jing Z, et al. (2012) Na+-taurocholate cotransporting polypeptide is a functional receptor for human Hepatitis B and D virus. Life.
- Huang HC, Chen CC, Chang WC, Tao MH. (20120 Entry of Hepatitis B virus into immortalized human primary hepatocytes by clathrin- independent endocytosis. J Virol. 86(17):9443-53.
- Herscher C, Pastor F, Burlaud-Gaillard J, Dumans A, Seigneuret F, et al. (2020) Hepatitis B virus entryinto HepG2- NTCP cells requires clathrin- mediated endocytosis . Cell Micro boil. 12:e13205.
- Iwamoto M, Saso W, Nishioka K, Ohki H, Sugiyama R, et al. (2020) Themachineryfor endocytosis of epidermal growth factor receptor coordinates the transport of the incoming Hepatitis B virus to the endosomal network. J Biol Chem. 295(3):800-7.
- Herscher C, Roingeard P, Blanchard B. (2020) Hepatitis B virus entry into cells. Cells.9(6):1486.
- Kock J, Nassal M, Deres K, Blum HE, von Weizsacker F. (2004) Hepatitis B virus nucleocapsid formed bycarboxy terminally mutated core proteins contain spliced viral genome but lack full size DNA . J Virol. 78(24):13812-8.
- Ludgate L, Liu K, Luckenbaugh L, Streck N, Enk S, et al. (2016) Cell free Hepatitis B virus capsids assembly dependent on the core protein C terminal domain and regulated by phosphorylation. J Virol. 90(12):5830-44.
- deRocquigne H, Rat V, Pastor F, DarlixJ L, Hourioux C, et al. (2020) Phosphorylation of the arginine rich C terminal domains of the Hepatitis B virus core protein as a fine regulator of the interaction between HBc and nucleic acid. Viruses. 12(7):738.
- Christie M, Chang CW, Rona G, Smith KM, Stewart AG, et al. (2016) Structural biology and regulation of proteins import into the nucleus. J Mol Biol. 428(10 Pt A):2060-90.
- Dohner K, Nagel CH, Sodeik B. (2005) Viral stop and go along microtubules: taking a ride dynein and kinesins. Trends Microbiol. 13(7):320-7.
- Rabe B, Glebe D, Kann M. (2006) Lipid mediated introduction of Hepatitis B virus capsids into non susceptible cells allows highly efficient replication and facilitates the study of early infection events. J Virol. 80(11):5465-73.
- Osseman Q, Gallucki L, Au S, Casenave C, Berdance E, et al. (2018) The chaperone dynein LL1 mediates cytoplasmic transport of empty and mature Hepatitis B virus capsids. J Hepatol. 68(3):441-8.
- Li HC, Huang EV, Su PV, Wu SY, Yang CC, et al. Nuclear export andimportof human Hepatitis B virus capsids proteins and particles.
- Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. (1999) Phosphorylation dependent binding of the Hepatitis B virus core particles to the nuclear pore complex. J Cell Biol. 145(1): 45-55.
- Guo H, Mao R, Block TM, Guo JT. (2003) Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J Virol. 84(1):387-96.
- Rabe B, Vlachou A, Pante N, Helenius A, Kann M. (2003) Nuclear import of human Hepatitis B virus capsids assembly and captures capsid dynamicsin vitro . Proc Natl Acad Sci USA.100:9849-54.
- Wang JC, Dhason MS, Zlotnik A. (2012) Structural organization of pregenomic RNA and the Carboxy-terminal domain of thecapsid protein of Hepatitis B virus. PLoS Pathogen. 8:e 1002919.
- Cui X, Ludgate L, Ning X, Hu J. (2013) Maturation associated destabilization of Hepatitis B virus nucleocapsid. J Virol. 87(21):11494-503.
- Hilmer JK, Zlotnik A, Bothner B. (2008) Conformational equilibra and rates of localized motion within Hepatitis B virus capsids. J Mol Biol. 375(2):581-94.
- Luo J, Xi J, Gao L, Hu J. (2020) Role of Hepatitis B virus capsids phosphorylation in nucleocapsid disassembly and covalently closed circular DNA formation. PLoS Pathogen. 16(3):e1008459.
- Chen C, Wang JC, Pierson E, Keifer DZ, Delaleau M, et al. (2016) importin beta can bind Hepatitis B virus core protein andempty core-like particles and induce structural changes. PLoS Pathogen. 12(8):e1005802
- Rabe B, Delaleau M, Bischof A, Foss M, Pumpens P, et al. (2009) Nuclear entry of Hepatitis B virus capsids involves disintegration to protein dimers followed by nuclear re-association to capsids. PLoS Pathogen. 5(8):e 1000563.
- Schmitz A, Schwarz A, Foss M, Zhou L, Rabe B, et al. (2010) Nucleosporins 153 arrests the nuclear import of Hepatitis B virus capsids in nuclear basket. PLoSPathogen 6(1):e1000741.
- Su PY, Yang CJ, Chu TJ, Chang CH, Chiang C, et al. (2016) HBV maintains electrostatic homeostasis modulating negative charges from phosphoserine and encapsidated nucleic acids. Sci Rep.6:38959.
- Cui X, Guo JT, Hu J. (2015) Hepatitis B virus covalently closed circular DNA formation in immortalized mouse hepatocytes associated with nucleocapsid destabilization. J Virol. 89(17):9021-28.
- Lempp FA, Mutz P, Lipps C, Wirth D, Bartenschlager R, et al. (2016) Evidence that Hepatitis B virus replication in mouse cells is limited by a lack of a host cell dependency factor. J Hepatol. 64(3):556-64.
- Duheron V, Nilles N, Pecenko S, Martinelli V, Fahrenkrog B. (2017) Localization of Nup 153 andSENP1to nuclear pore complexes is requiredfor 53BP1 mediated DNA doublestrand break repair. J Cell Sci. 130(14):2306-16.
- Lupberger J, Schedler S, Peiran A, Hildt E. (2013) Identification and characterization of a novel polymerase bipartite nuclear localization signal in the Hepatitis B virus polymerase. World J Gastroenterol. 19(44):8000-10.
- Kock J, Rosler C, Zhang JJ, Blum HE, Nassal M , et al. (2010) Generation of covalently closed circular DNA of Hepatitis B virus via intracellular recycling. PLoS Pathogen. 6(9):e1001082.
- Basagoundanavar SH, Perlman DH, Hu J. (2007) Regulation of hepadnavirus reverse transcription by dynamic nucleocapsid phosphorylation. J Virol. 81(4):1641-9.
- Verrier ER, Yim SA, Heydmann L, ElSaghire H, Bach C, et al. (2018) Hepatitis B virus evasion from cyclic guanosine monophosphate - adenosine monophosphate synthase sensing in human hepatocytes. Hepatology. 68(5):1695-09.
- Tsukuda S, Watashahi K. (2020) Hepatitis B virus biology and life cycle . Anti Virus Res. 182:104925.
- Koniger C, Wingert J, Marsmann M, Rosler C, Beck J, et al. (2014) Involvement of the host repair enzyme in TDP2 in formation of the virus covalently closed circular DNA persistence reservoir of Hepatitis B viruses. Proc Natl Acad Sci USA. 111(40):E4244- 53.
- Cai D,Yan R, Xu JZ, Zhang H, Shen S, et al. (2020) Characterization of the termini of cytoplasmic Hepatitis B virus deproteinated relaxed circular DNA. J Virol. 95(1):e00922-20.
- Long Q, Yan R, Hu J, Cai D, Mitra B, et al. (2017) The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathogen. 13(12):e1006784.
- Qi Y, Gao Z, Xu G, Feng B, Liu C, et al. (2016) DNA polymerase kappais a key cellular factor for the formation of covalently closed circular DNA of Hepatitis B virus. PLoS Pathogen. 12(10):e1005893.
- Wei L, Ploss A. (2020) Core components of DNA lagging strand synthesis machinery are essential for Hepatitis B virus ccc DNA formation. Nat Microbiol. 5(5):715-26.
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, et al. (2010) Three DNA polymerases, recruited by different molecular mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 37(5):714-27.
- Ogi T, Lehmamnn AR. (2006) The Y-family DNA polymerase kappa(pol kappa )functions in mammalian nucleotide-excision repair. Nat Cell Biol. 8(6):640-2.
- Tang L, Sheraz M, McGrane M, Chang J, Guo JT. (2019) DNA polymerase alpha is essential for intracellular amplification of Hepatitis B virus covalently closed circular DNA. PLoS Pathog. 15(4):e1007742.
- Kitamura K,Que L, Shimadu M, Koura M, Ishihara Y, et al. (2018) Flap endonuclease1 is involved in ccc DNA formation in the Hepatitis B virus. PLoS Pathog. 14(6):e1007124.
- Sheraz M, Cheng J, Tang L, Chang J, Guo JT. (2019) Sheraz Topoisomerases are required for the synthesis of Hepatitis B virus covalently closed circular DNA. J Virol. 93.
- Luo J, Cao V,Gao L, Hu J. (2017) Identification of an intermediate in Hepatitis B virus covalently closed circular(CCC) DNA detection. J Virol.91.
- Wei L, Ploss A. (2021) Hepatitis B virus cccDNA is formed through distinct repair processes of each strand. Nat Commun. 12(1):1591.
- Luo J, Luckenbaugh L, Hu H, Yan Z, Gao L, et al. (2020) Involvement of host ATR-CHK1 pathway in Hepatitis B virus covalently closed circular(CCC) DNA formation and sensitive and selective CCC DNAdetection.mBio. 11(1):e03423-19
- Fortini P, Doglioti E. (2010) Mechanisms of dealing with DNA damage in terminally differentiated cells. Mutat Res. 685(1-2):38-44.
- Gillmann R, Lopes Floro K, Wankell M, Hebbard L. (2021) The role of DNA damage and repair in liver Cancer . Biochim Biophys Acta Rev Cancer. 1875(1):188493.
- Nickoloff JA, Jones D, Lee SH, Williamson EA, Hromas R. (2017)Drugging the Cancers addicted to DNA repair. J Natl Cancer Inst. 109
- Bock T, Schwinn S, Locarnini S, Fyfe J, Manns MP, et al. (2001) Structural organizations of the Hepatitis B virus mini chromosome.J Mol Biol. 307(1):183-96.
- Tropberger P, Mercier A, Robinson M, Zhang W, Ganem DE, et al. (2015)Mapping of Histone modifications in episomal HBV ccc DNA uncovers an unusual chromatin organization amenable to Epigenetic manipulation. Proc Natl Acad Sci USA. 112(42):E5715- E5726.
- Belloni L, Pollicino T, De Nicuola F, Guerrieri F, Raffa G, et al. (2009) Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of ccc DNA function. Proc Natl Acad Sci U S A. 106(47):19975-9.
- Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrino G, et al. (2006) Hepatitis B virus replication is regulated by the acetylation status of Hepatitis B virus ccc DNA bound H3 and H4 Histones. Gastroenterology. 130(3):823-37.
- Riviere L, Gerossier L, Ducroux A, Dion S, Deng Q, et al. (2015) HBx relieves chromatin mediated transcriptional repressionof Hepatitis B virus ccc DNA involving SETDB1 Histone Methyltransferase. J Hepatol. 63(5):1093-1102.
- Flecken T, Meier MA, Skewes-Cox P, Barkan DT, Heim MH, et al. (2019) Mapping the heterogeneity Histone modificationson Hepatitis B HBV virus DNA using liver needle biopsies obtained from Chronically Infected patients. J Virol. 93(9):e02036-18.
- Alvarez-Astudillo F, Garrido D, VarasGodoy M, Gutierez JL, Villanueva RA, et al. (2021) The Histone variant H3.3 regulates the transcription of Hepatitis B virus. Ann Hepatol. 21:100261.
- Wang Z, Wang W, Wang L. (2020) Epigenetic regulation of covalently closed circular DNA mini chromosome in Hepatitis B virus infection. Biophys Rep. 66(6):115-26.
- Tu T, Budzinska MA, Shackel NA,Urban S. (2017) HBV DNA integration; molecular mechanisms and Clinical implications. Viruses. 9(4):75.
- Buendia MA, Neuveut C. (2015) Hepatocellular carcinoma. Cold Spring Harb Perspect Biol. 5:a021444.
- Zhang YY, Zhang BH, Theele D, Litwin S, Toll E, et al. (2003) Single cell of analysis of covalently closed circular DNA copy numbers in a hepadnavirus infected liver. Proc Natl Acad Sci U S A. 100(21):12372-7.
- Lucifora Z, Protzer U. (2016) Attacking Hepatitis B virus cccDNA:the holy grail of Hepatitis B cure. J Hepatol. 64(1):S41-S48.
- Boyd A, Lacombe K, Lavocat F, Maylin S, Miailhes P, et al. (2016) Decay of ccc DNA marks persistence of intrahepatic viral DNA synthesis under tenofovir inHIV-HBVcoinfected patients. J Hepatol. 65(4):683-91.
- Burdette D, Cathcart A, Shauf A, Win R, Zaboli S, et al. (2019) Evidence for the presence of infectious virus in the serum from Chronically Hepatitis B patients suppressed on nucleos(t)ide therapy withdetectable but not quantifiable HBV DNA. J Hepatol. 70(1):e95.
- Huang Q, Zhou B, Cai D, Zong Y, Wu Y, et al. (2021) Rapid turnover of Hepatitis B viruscovalently closed circular DNAindicated by monitoring emergence and reversion of signature-mutation in treated Chronic Hepatitis B patients. Hepatology. 73(1):41-52.
- Alexopoulou A, Vasiilieva L, Karayiannis P. (2020) New approaches to the treatment of Chronic Hepatitis B. J Clin Med. 9(10):3187.
- Yang L, Liu F, Tong X, Hoffmann D, Zou J,et al. (2019) Treatment of Chronic Hepatitis B virus infection using small molecular modulators of nucleocapsids assembly: recent advances and perspectives. ACS Infect Dis. 5:713-24.
- Zheng Y, Yang L, Yu L, Zhu Y, Wu Y, et al. (2023) Canocapaviris a novel capsid assembly modulators inducing a conformational change of the linker Region of HBV core protein. Viruses. 15(5):1195.
- Lee HW, Lee JS, Ahn SH. (2020) Hepatitis B virus cure: targets and future therapies. Int J Mol Sci. 22(1):213.
- Tang L, An P, Zhao Q, Winkler CA, Chang J, et al. (2023) Heat shock protein family a member1promotes intracellular amplification of covalently closed circular DNA. J Virol. 97(1):e0226122.
- Mendelhall M, Hong X, Hu J. (2023) Hepatitis B virus capsid: the core in productive entry and covalently closed circular DNA formation. Viruses. 15(3):642.
- Hong X, Bianchini EN, Wang JCN, Hu J. (2023) Characterization of intracellular pre-core derived protein and their function in Hepatitis B virus infected hepatocytes . mBio. 14(1):e0350122.
- Xi J,Cui X, Liu K, Liu H, Wang J, et al. (2022) Region specific Hepatitis B virus virus genome infection exposure fromnucleocapsids modulated bycapsidlinkersequenc and inhibitor: implications for uncoating. J Virol. 96(8):e0039922.
- Anderson M, Stac M, Thi EP, Pichhio G, Mbanya D, et al. (20203) Measuring Hepatitis B pg RNA stability using an updated automated HBV B pg RNA with increased sensitivity. Hepatol Commun. 7(4):e0099

