The Impact of Sequencing Human Genome on Genetically Inherited Diseases
Hameed Khan*
Department of Genetics & Robotics, Senior Scientist, NCMRR (National Center for Medical Rehabilitation Research), National Institutes of Health (NIH), Adjunct Professor NYLF, Bethesda, Maryland, USA
*Corresponding author: Hameed Khan, Department of Genetics & Robotics, Senior Scientist, NCMRR (National Center for Medical Rehabilitation Research), National Institutes of Health (NIH), Adjunct Professor NYLF, Bethesda, Maryland, USA
Citation: Khan H. (2023) The Impact of Sequencing Human Genome on Genetically Inherited Diseases. J Can Ther Res. 3(1):1-26.
Received: November 23, 2022 | Published: January 11, 2023
Copyright© 2023 Genesis Pub by Khan H. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International License (CC BY 4.0). This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are properly credited.
DOI: https://doi.org/10.52793/JCTR.2023.3(1)-21
Abstract
This abstract attempts to explore if genetically inherited diseases could be prevented before conception. Sequencing Egg and Sperm before conception and comparing with the Reference Sequence will identify any deleterious mutations which will predict the onset of any serious illness in the embryo. Male and female differ genetically by their respective sex chromosome composition, that is, XY as male and XX as female. Although both X and Y chromosomes evolved from the same ancestor pair of auto somes, the Y chromosome harbors male-specific genes, which play pivotal roles in male sex determination, germ cell differentiation, and masculinization of various tissues. While the X-chromosome of the mother is made of 164 million AT-GC nucleotide base pairs and carries 1,144 genes, the Y-chromosome of the father is made of 59 million A-T-GC base pairs and carries 231 genes. The next generation of Nanopore Sequencer will sequence the X and Y-chromosome cheaper, faster and with great precision and accuracy. To this date, more than 6,000 genetic disorders are identified, and new genetic disorder are constantly being added in the medical literature. Some of the confirmed disorders include. Down syndrome (Trisomy 21), Fragile X syndrome, Klinefelter syndrome, Turner syndrome, Cystic Fibrosis, Thalassemia., Sickle Cell Anemia, Huntington's Disease, Duchenne's Muscular Dystrophy, Tay-Sachs Disease etc. Although some of these genetic diseases are treatable, Most of them remain under investigation. Once a mutation related to a disease is identified, our next challenge is to design drugs to shut of that mutated gene. Since the language of life Aziridine, 2-4, Dinitrophenyl Benzamide(CB1954) to treat a solid aggressive tumor like the Walker carcinoma 256 in Rats and how similar rational was used to design Di-aziridine benzoquinoneCarbamate (AZQ)(US Patent 4,233,215) to treat solid aggressive brain tumor like Glioblastoma in humans.
Keywords
Sequencing Genome; Mutations; Genetic Diversity; Genome diseases; Inbreeding; AZQ
A Note to my readers
The Impact of Sequencing Human Genomes are a series of lectures to be delivered to the scholars of the National Youth League Forum (NYLF) and the International Science Conferences. NYLF scholars are the very best and brightest students selected from all over the USA and the world brought to Washington by Envision, an outstanding organization that provides future leaders of the world. I am reproducing here part of the lecture which was delivered at the International Science Conference that was PCS 6the Annual Global Cancer Conference held on November 15-16, 2019, in Athens, Greece.
Special notes
I am describing below the use of highly toxic lethal chemical weapons (Nitrogen Mustard) which was used during WWI and its more toxic analogs developed as more toxic weapons during WWII. I described the use of Nitrogen Mustard as anti-cancer agents in a semi-autographical way to accept the responsibility of its use. When we publish research papers, we share the glory with colleagues and use the pronoun “We” but only when we share the glory not the misery. In this article by adding the names of my coworkers, the animal handers, I will share only misery. The Safety Committee is interested to know who generated the highly lethal Chemical Waste, How much was it generated and how was it disposed. I accept the responsibility. The article below sounds semi-auto-biographical, it is, because I am alone responsible for making these compounds of Nitrogen Mustard, Aziridines and Carbamate. To get a five-gram sample for animal screening, I must start with 80 grams of initial chemicals for a four-step synthesis. To avoid generating too much toxic chemical waste, instead of using one experiment with 80 grams, I conducted 80 experiments with one gram sample, isolating one crystal of the final product at a time. The tiny amount of waste generated at each experiment was burned and buried at a safe place according to safety committee rules.
Introduction
At the dawn of new millennium, we embarked on three great revolutions in science. A Quantum Revolution, a Computer Revolution, and a Genetic Revolution. These revolutions deal with matter, mind, and life.
Quantom Revolution
The first is the Quantum revolution. In Quantum Revolution, we solved the mystery of matter. We found that total matter in the Universe is made of atoms. The quantum revolution allows us to understandthe world of the very small and the fundamental properties of the matter. Our deepest understanding of the atomic world comes from the advent of the quantum theory. Our Universe is made of matter and matter is made of atoms. To understand the Universe, we study a single atom; we found that the atom is made of a positively charged Proton with Neutron in the central nucleus around which revolve the negatively charged electrons in a specific orbits carrying a unit amount of energy called the quantum energy. It is the nucleus that carries tremendous amount of energy. In fission reactions when we break the nucleus, we could release that energy? The nucleus is made of positively charged protons and neutral neutrons. Since positively charged protons repel each other, they are fused together by nuclear forces that hold the protons tightly together. When Hydrogen atoms fused together, they form Helium atoms releasing high energy neutron which travels at a speed of about 10 million meters per second or about three percent of the speed of light. The highly charged neutrons bombard a radioactive atom like Uranium atom, it undergoes fission, which releases energy and several more neutrons, which then split more Uranium atoms and release more energy and neutrons.This sets a nuclear chain reaction. A chain reaction refers to a process in which neutrons released in fission produce an additional fission in at least one further nucleus. This nucleus in turn produces more neutrons, and the process repeats until all atoms are broken releasing tremendous amount of energyat once. With a ten-poundpure Urainium-235, we could generate so much energy that a city like Hiroshima can be turned to ashes in minutes. Or if we control the chain-reaction by placing a rod of Boron or Cadmium which absorbs neutrons to control the chain-reaction, we could convert matter into the nuclear energy that could be converted into electricity. The process may be controlled (nuclear power) or uncontrolled (nuclear weapons).
Our deepest understanding of the atomic world comes from the advent of the quantum theory. It is the quantum revolution nearly a century ago that led to the 20th-century technological revolution, which was based on the transistor, laser, and atomic clock. These inventions gave us computers, optical fiber communication, cell phones and the global positioning system, all of which are vital to the world economy. The greatest achievement we made in Quantum revolution is that we learned to convert matter into energy. Energy that runs the engine of modern society.
Computer revolution
The second is the computer revolution. It was the revolution of mind. It brought the information age. Using a binary code (zero and one), we wrote programs to store all the information we generated from the dawn of human civilization to the present day. We can not only store information, but also, we could retrieve, cut, and paste and move the information around with the speed of light. Our greatest accomplishment is that we have capture space time. We can communicate with anyone around the world within seven second.
Genetic revolution
The third is the genetic revolution, the greatest revolution of them all. It solves the mystery of life. Now, we know with fair certainty how life could have evolved at some remote corner of the Earth about three and a half billion years ago and how it crawled on evolutionary path to fly at will or stay still. In a three-billion-year journey across time, it reached us and help us develop our language our consciousness and our mind. We are the most intelligent of all living creatures on Earth. We are so intelligent that we ask questions about ourselves who we are and where have we all come from? What was it that made us this way? While hundreds of experiments were conducted to answer the first two questions, a single experiment answers the last question. It was the greatest experiment ever conceived by human mind. It will answer the most fundamental question we have asked ourselves since the dawn of human civilization; Who are we? Where have we all have come from and what was it that made us this way. The answers to these questions are embedded in the nucleus of every cell of our body. Our book of life is written in the nucleus of each cell. Reading the total genetic information that makes us is called the Human Genome. We decided to read our entire genome letter by letter, word by word and sentence by sentence under a project entitled The Human Genome Project. The goal of the human genome project is to determine the total number of base pairs nucleotides and the order in which they are arranged that make us human. A string of nucleotides called the De oxyribose Nucleic Acid(DNA) and identifying how DNA got together to make genes and mapping, and sequencing all of the genes of the human genome from both a physical and a functional standpoint.
The Sequencing of the Human Genome which is not only reading the entire book of life of human being letter, by letter, word by word and sentence by sentence, chapter by chapter but also the order in which these letters are arranged called sequencing, is the greatest discovery of all times. The sequencing of the Human Genome will answer the most fundamental questions, we have asked ourselves since the dawn of human civilization; what it means to be human; what the nature of our memory and our conscientiousness is; our development from a single cell to a complete human being; the biochemical basis of our senses; the process of our aging; the scientific basis of our similarity and dissimilarity. Similarities that all living creatures from a tiny blade of grass to the mighty Elephants including man, mouse, monkey, mosquitos, and microbes are all made of the same chemical building blocks, the nucleotides, and yet we are so diverse that no two individuals are alike, even identical twins are not exactly identical; they grow up to become two separate individuals.
To understand the basis of all diseases, we must read and understand the total genetic information that makes us humans that is to read the genome of a healthy human being (called the Reference Sequence) and compare with the sick person’s genome to identify the spelling errors in the genome responsible for causing the disease. That is how the story of our book of life begins: As we all know that we are the loving union of our parents. Our mother’s egg receives our father’s sperm, and we are conceived. The fertilized egg carries complete information to make us. More than seventy years ago, the Nobel Laureate, Irvin Schrödinger, was the first person to propose that the hereditary molecule must contain a "code-script" that determined "the entire pattern of the individual's future development and of its functioning in the mature state". This was the first clear suggestion that genes contained some kind of "code". Now, we know that the essence of life is information and genes are the bearers of that information, carrying it in a tiny, complex code inside every cell of our bodies. If we examine for comparison, the fertilized egg of a human, mouse, and monkey under a microscope. We observed that all fertilized eggs look the same and yet first fertilized egg carries the instructions to make a man, the second carries the information to make a mouse and third carries the information to make a monkey. We are certain that there exists a secret code within those fertilized eggs to make different species; Schrodinger called that secret code, the Script Code, now known as the Genetic Code. To understand the secret code, we must examine the internal structure of the fertilized egg. We propose that we examine three “C”. The first C stands for the chromosome, the coloring bodies present inside each cell. The traits to make man, mouse or monkey must be located on the Chromosomes. These traits must be held together tightly by the second C, the covalent bonds. As the living cells grow, they must have the ability to copy the instructions accurately that copying is the third C. The Genetic Code to make man, mouse or monkey must be written on the chromosomes. Based on this information Crick and Watson broke the Genetic Code and unlocked the secret of life. If we unlock the secret of life, we will understand how evolution puts the traits together over millennia to separate man, from mouse and mouse from monkey. By unlocking the secret of life, we can understand how the normal cells work and how the normal cells become abnormal leading to all diseases including cancerous.
On further examination, we found that the chromosomes are made of four chemicals and information is located on them and these four molecules are called nucleotides bases. These bases are made of Deoxy Ribonucleic Acid (DNA). DNA is made of a string of nucleotides. It is a storehouse of information and is made of the same four nucleotide bases and they are: Adenine (A), Thymine (T), Guanine (G), and Cytosine (C). According to Crick’s Central Dogma, [1] the information flows from the DNA which is transcribed into RNA which is translated in Ribosome into proteins. RNA is converted into an active form and is transcribed into mRNA (or messenger RNA after splicing out noncoding DNA) and by converting Thymine to active form Uracil (U) and from a double stranded DNA to a single stranded RNA and where the sugar Deoxy Ribose is replaced by sugar Ribose. The mRNA is translated by Ribosome into proteins. Gene Expression begins in Ribosome when a 4-letter genetic text is converted to a three-letter Codon which code for a single amino acid. By comparing Gene Profiles of normal genes with mutated genes, one can identify with precision and accuracy the exact location of mutated (altered or damage) nucleotide responsible for causing the disease. Comparing Gene Profiles is an excellent diagnostic method which helps us design drugs to specifically shut off the mutated genes.
Seventy years ago, Schrödinger predicted secret code of life using such a poor resolution microscope that we don’t even use in our high school today. Instead, we use electron microscope. We can magnify the same fertilized egg to a million times of its original size, almost the size of a house. What we observe inside the fertilized egg is very analogous to the house. The house has a kitchen; the cell has a nucleus. Suppose your kitchen has a shelf which contains 46 volumes of cookbooks which contain 24,000 recipes which carry instructions to cook food for your breakfast, lunch, and dinner. The nucleus in the fertilized egg contains 46 chromosomes; (23 from our mother and 23 from our father), which carry 24,000 chapters called genes. Genes are units of inheritance which code for all 20 amino acids. Hundreds of amino acids join to form a protein and thousands of proteins interact to make a cell. Millions of cells interact to make an organ and several organs interact to make a man or a mouse or a monkey. The number and the order of the nucleotides determine the composition of a species. [2-3].
If the cookbook in your kitchen is written in English language, it uses 26 letters, but the book of life of all living creatures is written in 4 letters and they are A, T, G and C. These are the initials of four chemicals called nucleotides found the nucleus of all living cells. Nucleotides are made of sugar Ribose (Deoxy Ribose in DNA and Ribose in RNA), a phosphate group and one of the four Nitrogen bases, two Purines and two Pyrimidines and the Thymine is converted to Uracil in RNA. These molecules are found in the nucleus of all living cells from a tiny blade of Grass to mighty elephant including man, mouse, and monkey. The total genetic information to make any living creature is based on the above four-letter text and out of these four letters, only three letter Codon which carries the Genetic Code for an amino acid (such as GUU is for amino acid Valine, GCU is for Alanine, GAA is for Glutamine etc.) the building blocks for all proteins. Sixty-four codons code for 20 amino acids and codons for all 20 amino acids have been decoded. All living creatures use the same genetic code. A string of these nucleotides is called the DNA (Deoxy Ribonucleic Acid). Reading the number and the order of nucleotides are called genome sequencing[4-5].
As I said above, a gene is a piece of DNA. Out of four nucleotides text, three letters code for an amino acid called codon. A gene is made of several hundred codons. On a piece of DNA, a gene is identified by a start and a stop codon. The start codon is AUG which codes for the amino acid Methionine and there are three stop codons, and they are UAG, UGG, and UGA. The extension of DNA synthesis stops once the one of the three stop codons appears. A gene codes for a protein. We found that the smallest gene in bacterial genome and the largest gene is in Duchenne Muscular Dystrophy genome. To code for a protein, between start and stop codon, a gene has accumulated several hundreds to several thousand codons. A single mutation in the coding region of a single codon will alter the gene function. Mutation is caused by either exposure to radiations, chemical or environmental pollution, genetic inheritance, viral infections or DNA deletion, insertion relocation or inversion. Mutation can be good, bad, or neutral. A good mutation can convert a single cell organism to a multicellular creature resulting in evolution. A bad mutation is responsible for coding for a wrong amino acid responsible for causing diseases. A neutral mutation can serve as a gene marker identifying its presence close to a good or a bad gene. By sequencing Human Genome, we have identified 16,000 good genes, 6,000 bad genes and 2,000 pseudo or neutral genes. Less than 2% of our genome codes for proteins. The remaining 98% of our genome carry pieces of DNA which serves as switches, promotors, enhancers etc. The greatest Darwinian transformation is controlled by switches. By switching on and off a gene, the body plan gene called the FOX gene can bring the evolutionary changes in the body. Genes code for protein, it is the switches that turn genes on or off.
Gene Expression begins in Ribosome when a 4-letter genetic text is converted to a three-letter Codon. By comparing Gene Profiles of normal genes with mutated genes, one can identify with precision and accuracy the exact location of mutated (altered or damage) nucleotide responsible for causing the diseases. Comparing Gene Profiles is an excellent diagnostic method which helps us design drugs to specifically shut off the mutated genes. Delivering drugs from injection site to the target site is the essential way of treating diseases. We cannot design novel drugs unless we find the abnormal mutations responsible for causing that disease. The reading of the total genetic information that make us human is called the Human Genome. The reading the entire book of our life is authorized by the US Congress under The Human Genome Project.
In 1990, US Congress authorized three billion dollars to NIH to decipher the entire Human Genome under the title, “The Human Genome Project.” We found that our genome contains six billion four hundred million nucleotides base pairs half comes from our father and another half comes from our mother. Less than two percent of our Genome contains genes which code for proteins. The other 98 percent of our genome contains switches, promoters, terminators etc. The 46 Chromosomes present in each cell of our body are the greatest library of the Human Book of Life on planet Earth. The Chromosomes carry genes which are written in nucleotides base pairs. Before sequencing (determining the number and the order of the four nucleotides on a Chromosomes), it is essential to know how many genes are present on each Chromosome in our Genome. The Human Genome Project has identified not only the number of nucleotide base pairs on each Chromosome, but also the number of genes on each chromosome [6].
The following list provide the details composition of each Chromosome including the number of nucleotides and the number of genes on each Chromosome:
We found that the Chromosome-1 is the largest Chromosome carrying 263 million A, T, G and C nucleotides bases and it has only 2,610 genes. The Chromosome-2 contains 255 million nucleotides bases and has only 1,748 genes. The Chromosome-3 contains 214 million nucleotide bases and carries 1,381 genes. The Chromosome-4 contains 203 million nucleotide bases and carries 1,024 genes. The Chromosome-5 contains 194 million nucleotide bases and carries 1,190 genes. The Chromosome-6 contains 183 million nucleotide bases and carries 1,394 genes. The Chromosome-7 contains 171 million nucleotide bases and carries 1,378 genes. The Chromosome-8 contains 155 million nucleotide bases and carries 927 genes. The Chromosome-9 contains 145 million nucleotide bases and carries 1,076 genes. The Chromosome-10 contains 144 million nucleotide bases and carries 983 genes. The Chromosome-11 contains 144 million nucleotide bases and carries 1,692 genes. The Chromosome-12 contains 143 million nucleotide bases and carries 1,268 genes. The Chromosome-13 contains 114 million nucleotide bases and carries 496 genes. The Chromosome-14 contains 109 million nucleotide bases and carries 1,173 genes. The Chromosome-15 contains 106 million nucleotide bases and carries 906 genes. The Chromosome-16 contains 98 million nucleotide bases and carries 1,032 genes. The Chromosome-17 contains 92 million nucleotide bases and carries 1,394 genes. The Chromosome-18 contains 85 million nucleotide bases and carries 400 genes. The Chromosome-19 contains 67 million nucleotide bases and carries 1,592 genes. The Chromosome-20 contains 72 million nucleotide bases and carries 710 genes. The Chromosome-21 contains 50 million nucleotide bases and carries 337 genes. Chromosome-22 contains 56 million nucleotides and carries 701 genes. Finally, the sex chromosome of all females called the (X) contains 164 million nucleotide bases and carries 1,141 genes. The male sperm chromosome contains 59 million nucleotide bases and carries 255 genes.
If you add up all genes in the 23 pairs of Chromosomes, they come up to 26,808 genes and yet we keep on mentioning 24,000 genes needed to keep us function normally. Out of 24,000 genes, we have identified 16,000 good genes, 6,000 bad genes and 2,000 Pseudo genes. As I said above, a gene codes for a protein, not all 24,000 genes code for proteins. It is estimated that less than 19,000 genes code for protein. Because of the alternative splicing, each gene codes for more than one protein. All functional genes in our body make less than 50,000 protein which interact in millions of different ways to give a single cell. Millions of cells interact to give a tissue, hundreds of tissues interact to give an organ and several organs interact to make a human.
Not all genes act simultaneously to make us function normally. Current studies show that a minimum of 2,000 genes are enough to keep human function normally; the remaining genes are backup support system, and they are used when needed. The non-functional genes are called the Pseudo genes. For example, millions of years ago, humans and dogs shared some of the same ancestral genes; we both carry the same olfactory genes, only in dogs they still function to search for food. Since humans don’t use these genes to smell for searching food, these genes are broken and lost their functions, but we still carry them. We call them Pseudo genes. Recently, some Japanese scientists have activated the Pseudo genes, this work may create ethical problem in future as more and more Pseudo genes are activated. Nature has good reasons to shut off those Pseudo genes.
On April 3, 2003, we sequenced the entire Human Genome. We not only read the entire script of our genome, letter by letter, word by word, sentence by sentence, but also we read the number of letters and the order in which they are arranged (sequence) called under the title, “The Human Genome Project”. We found that less than two percent of the Gene in our Genome codes for proteins and the rest is the non-coding regions which contains switches to turn the genes On or Off, pieces of DNA which act as promoters and enhancers of the genes. Using restriction enzymes (which act as molecular scissors), we can cut, paste, and copy genetic letters in the non-coding region which could serve as markers and which has no effect, but a slight change in the coding region makes a normal cell become abnormal or cancerous. Recent studies showed that mutations in switches, promotors and enhancers which are present in the non-coding regions are also responsible for some unusual diseases. We need to go back and look at these regions more carefully.
Our Genome provides the genetic road map of all our genes, past, present and future. For example, it can tell us how many good or bad genes we inherit from our parents and how many of those gene we are going to pass on to our children. If a family has too many bad genes, and have a family history of serious illnesses, you can break off the information flow and stop having children or stop donating mutated eggs and sperms.
Our search for unknown diseases has come to a closure:
There are two most powerful implications of the human Genome Sequencing. One of them is that we have come to closure. What it means is that we have the catalog of all genes in the Human Genome, we can search the entire genome and locate the desired gene. we will not wonder in the wilderness anymore. Everything there is to know about human health and traits are written on these genes in nucleotide sequences. Our Genomes provides the catalog of all genes.
We can scan the whole genome (Reference Sequence) for its response to a given situation. When we look at a normal cell and compare with an abnormal cell, we see the differences. Or when we compare their gene expression looking for a specific proteins, from a specific genes and for a specific nucleotide sequence, we can identify a specific disease. In the olden days, before sequencing human genome, a physician would order several tests and would say to his patient, I don’t know what is wrong with you, I will see if any of these tests show if my guess is right and if it is wrong, he will recommend few more tests to see if he could identify the illness. The guesswork and the trial-and-error days are over. Now, after sequencing the human genome, the physician would say I don’t know what is wrong with you, but I do know where to find it. It is written in your Genome. It would be easy for a Physician to scan the patient entire genome and compare against the Reference Sequence to identify the mutations responsible for causing the disease. We will take a small blood sample of the patient, separate his WBC, extract DNA, sequence his Genome and compare with the Reference Sequence letter by letter, word by word by word and sentence by sentence, we can easily identify the mutations responsible for causing the disease. The result will provide the best diagnostic method to identify a disease.
Our Genome is not just a diagnostic road map of our genes, it also tells us to clone the good genes and shut off the bad genes. Using the good genes, it also tells us to make its large-scale protein for worldwide use such as Insulin and Human growth hormone. On the other hand, identifying the bad genes and tell us to design drugs to shut off bad genes responsible for causing Cancers, Cardiovascular disease, and Alzheimer. We have already demonstrated that using the genetic engineering techniques, we can cut, paste, copy, and sequence a good gene for industrial scale preparation such as Insulin to treat 300 millions of diabetic around the world. Similarly Human Growth Hormone, once available in minute quantities from the pituitary glands of humans’ cadaver can now be produced in large amount in the Labs using the same genetic engineering method. Many valuable medicines are producedincluding Interleukin-II, for the treatment of the Kidney cancer, Factor-VIII for treating Hemophilia, Hepatitis B vaccine, Eleuthero protein for anemia, Whooping cough vaccine, and for Somatotropin for treating dwarfism.
Once the good and bad genes are identified, we know that the good genes code for good proteins which keep us healthy, and the bad genes produce bad protein that make us sick. Genome sequencing of bad genes start a new era of Genomic Medicine which is based on the genetic make-up of the individuals. The next step would be to design drugs to shut off the mutated genes. Gene Therapy will work if the disease is caused by a single gene mutation. Drug Therapy will work if multiple genes are responsible for causing diseases such as Cancers, Cardiovascular diseases, and Alzheimer.
Genetic Engineering and Recombinant Technology
It involves cutting, pasting, copying, and sequencing DNA from the genome. As I said above, in humans, less than two percent of the Genome codes for proteins and the remaining 98 percent of Genome contains non-coding regions which carry switches, enhancers, promoters, inhibitors etc. Only two percent of the Genome transcribes into RNA. The non-coding regions of the RNA is spliced out into mRNA which carry three letter Codons. Each codon codes for a single amino acid except in the alternative splicing. It is the Codons which is translated in the Ribosome into proteins. The protein carries out our body functions as soon as it folds and becomes three dimensional. As the Central Dogma of Francis Crick describes that double stranded DNA replicates (makes its own copies) in the nucleus and it transcribes into the single stranded RNA as it leaves nucleus as mRNA in the cytoplasm (after splicing out non-coding sequence) which is translated in the Ribosomes into proteins. Information flows from both Good genes and bad genes from nucleus into the cell keeping the organism healthy or sick. Good proteins from good genes keep us healthy and bad proteins from mutated genes produce bad proteins that make us sick. The flow of information is continuous and uninterrupted. Information flows from both Good genes and bad genes from nucleus into the cell keeping the organism healthy or sick. Good proteins from good genes keep us healthy and bad proteins from mutated genes produce bad proteins that make us sick.
One of the greatest challenges of the 21st century medicine is to cut out and purify a single gene from the string of genes from the entire human genome and insert this single human gene into a biological system making a recombinant fragment called Vector. Vectors protect genes from enzymatic destruction. Many microbial life form could serve as Vectors (such as bacteria or plasmids for smaller genes, and for larger genes using Phagemids, or Cosmids (Cosmid vectors are hybrids between plasmid and phage λ vectors. Cosmid vectors are designed to clone large fragments of DNA and to grow their DNA as a virus or as a plasmid. Cosmid. Vectors are used in homologous recombination between two different plasmids in the same cell. To scale up, the Vectors are harvested in Yeast or in bacteria. Later, large number of pure genes are isolated by breaking the Vectors with restriction enzymes.
Genes that carry instructions to made proteins are expressed in a two-step process and they are Transcription and Translation. First, theamino acids Codons are spliced or inserted into a double stranded DNA which is later transcribed into a single stranded m-RNA by removing non-coding nucleotides. As I said above, it is the m-RNA which is translated in the Ribosomes into all 20 amino acids. The Cells decode m-RNA in groups of three nucleotides called Codons which carry instructions to produce the amino acids. It is also worth repeating that when double stranded DNA is transcribed into a single stranded m-RNA, the nucleotide Thiamin is converted to Uracil. The Methyl group of Thiamine is replaced by a more water- soluble Hydroxyl group forming the Uracil. The nucleotide T for Thiamin is replaced by U for the Uracil. The m-RNA is translated into amino acids in Ribosomes. The gene expression has a Start Codon (AUG) which codes for amino acid Methionine and there are three Stop Codon which are UGG, UAG and UGA. Once the Stop Codon appears at the tail end of the DNA, amino acids synthesis stops. The Codons for each essential amino acid and their alternative codons are described below:
Valine (GTT, GTC, GTA, GTG), Leucine (CTT, CTC, CTA, CTG; TTA, TTG), Isoleucine (ATT, ATC, ATA), Phenylalanine (TTT, TTC), Tryptophan (TGG), Lysine (AAA, AAG), arginine (CGT, CGC, CGA, CGG; AGA, AGG), Histidine (CAT, CAC), Methionine (ATG), Threonine (ACT, ACC, ACA, ACG). In the past, all that repetition made it impossible to assemble some chopped-up pieces of DNA in the correct order. It’s like having identical puzzle pieces – scientists didn’t know which went were, leaving big gaps in the genomic picture. Now, with arrival of new sequencers, we read the book of our life cheaper and faster. The key advances included rapid improvements in the next generation gene sequencing machines made by Oxford Nanopore Technologies and Pacific Biosciences. The new Nanopore machine can accurately read a million letters of DNA at a time. Pacific Biosciences introduced a new sequencing machine which generate long-read sequencing that could read DNA sequence with greater than 99 percent accuracy. As the cost of sequencing goes down, all of us should have our genome sequenced. This genetic profile will serve as a part of our medical record. It will provide instant medical help.
Genomic Medicine
(What to do with good genes? cut, paste, and copy the gene)
Out of 24 thousand genes in our genome, we carry sixteen thousand good genes; six thousand bad genes or mutated genes which are responsible for six thousand diseases and two thousand Pseudogenes which are broken and have lost their functions. We also learned that about a minimum of 2,000 genes are essential to maintain a life and the remaining genes are backup or supporting genes to provide immediate help if needed. Using the Restriction Enzymes, like EcoR1, we could cut out all good and bad genes from the human genome to sequence and clone them by PCR. We need to study bad genes as well as good genes to design drugs to shut off defected genes. Diseases are caused by a single defected gene (calledMendelian disorders, for example, occur when specific mutations in single genes — called germline mutations — are inherited from either of one's two parents. Well-known examples of single genetic defects called the Mendelian diseases include cystic fibrosis, sickle cell disease, and Duchenne muscular dystrophyor multiple genetic defects (such as Cancers, Cardiovascular diseases or Alzheimer).
Gene Therapy
In Gene Therapy a single bad gene is replaced by a good gene. With single genetic defects, we could cut, paste, and copy good gene in a Vector such as flu virus and infect patients’ WBC with the transgenic virus which replace bad gene with the good gene. In a single genetic defect, Gene Therapy works supremely well as in the treatment of SCID (Severe Combined Immune Deficiency Syndrome) which is caused by a single genetic defect.
The supremely successful example of Gene Therapy is the treatment of Severe Combined Immune Deficiency Syndrome (SCID). SCID is also known as the Bubble Boy Syndrome. Because of a single genetic defect. patients lack the ability to fight off environmental microbes. To protect them from any infection, they are provided purified air in an enclosed environment. French Anderson and Mike Blaise at our Institute (NIH), using genetic engineering technique replaced the defected gene with a healthy gene. After harvesting in a harmless virus, infected the WBC obtained from the patient. The trans-genic WBC in vitro, returned the modified WBC to the same patient. Patients recovered and lives a normal life. With multiple genetic defects such as in cancers, Gene Therapy will not work, but Drug Therapy will work. We could design drugs to shut off multiple genes and prevent it from producing bad protein or cancers.
The first step is to cut the human genome at a specific site with a specific enzyme (prepare a Restriction Site Map) at the specific sites using restriction enzymes (which serves as molecular scissors such as EcoR1) first accomplished by El Salvador Luria, Max Delbruck, and Hamilton Smith. The fragment of human DNA (a single gene) if not protected will be destroyed by antibody. A naked gene is a piece of DNA (which has a start codon AUG and after a few thousand nucleotide (codons) end at one of the three stop codons UAG, UGA or UGG). If a gene is not protected by recombinant technology (making a hybrid) that is by recombining with the DNA of Virus, or Plasmids, or Chloroplasts (for plants) which serves as Vectors, will be destroyed by enzymes. One can store the fragments or genes in the Vectors once the human DNA fragment is stabilized in Vectors by recombinant technology; we can not only purify this fragment (genes), but also, we can make millions of copies of this fragment of DNA by transferring into the host cells such as Bacteria, mammalian cells or Yeast cell which autonomously replicates to produce library of genes. Each Library contains millions of copies of identical genes that produce same protein. Before the genetic revolution, Insulin is extracted from pancreas of the slaughtered animals which is used to treat old diseases such as diabetes; a tiny fragment of impurity could set anaphylactic shock and kill the patients. Now, highly pure human Insulin is produced by Genetic Engineering Technique and is used to treat 300 million diabetic patients worldwide without the loss of a single life using the same recombinant technology. Other products of Genomic Medicine such as Growth hormones and hormone proteins to treat Hemophilia by factor VIII protein are being developed as genomic medicines by recombinant technology.
Drug Therapy
(What to do with bad Genes? Either to replace by Gene Therapy or to design drugs to shut off bad genes).
The essence of life is information, and the information is located on the four nucleotide bases A-T and G-C. According to Central Dogma of Crick and Watson, the information on DNA is transcribed on RNA which is translated in Ribosome to protein. Attempts are being made to design drugs to attack cancer cells on all three level that is DNA, RNA and Protein. Herceptin, a novel class of drug, has been successful in attacking protein. Craig Milo has designed double stranded RNA to shut off gene and prevents its translation into protein. Attack on DNA to shut off a gene presents the greatest challenge because of the number of mutations on cancer cells.
How to design drugs to shut off bad gene variants?
We design drugs to shut off genes responsible for causing cancers because largest amount of funds is available to the National Cancer Institute (NCI) about $5B per year. Cancer is the leading cause of death and has surpassed the death of cardiovascular diseases. Over 636,000 people died of cancer; 1.9 million new cases will be diagnosed this year including 78,000 Prostate Cancer, 40,000 Breast cancer, 16000 Lung and Bronchus Cancer and 15,000 Colon and Rectal Cancer. Once diagnosed mutation by Gene Sequencing, the next step is to design drug to shut off those genes. I describe below my own work how I designed drugs to shut off cancer causing genes in animals and then translated the work in humans. (see special note-II above).
The rational drug design to treat cancers
All three old age diseases that is Cancer, Cardiovascular Diseases and Alzheimer carry multiple mutated genes responsible for causing these diseases. In each of the above three diseases, it is the harmful mutated genes that code for wrong protein which causes these diseases. If we design drugs to shut off mutated genes in one disease, using the same rationale, we should be able to shut off bad genes in all three old age diseases. Although Coronary Artery disease is a complex disease, researchers have found about 60 genomic variants that are present more frequently in people with coronary artery disease. Most of these variants are dispersed across the genome and do not cluster at one specific chromosome. During WWI, highly toxic chemicals are used as chemical weapons. These chemicals attack the DNA shutting off gene. After the war, scientists modified these chemicals to shut off malignant genes to treat diseases. To reduce toxicity, prodrugs are designed to seek out the specific malignant genes which replicate faster producing acids. Aziridines and Carbamate moieties are prodrugs, stable in neutral and basic media, but sensitive to acid. Drugs carrying Aziridines, and Carbamate moieties are broken down in acidic media generating Carbonium ions which attack DNA shutting off genes. Only the acid producing genes will be attacked no matter where they are located. It does not matter whether they are clustered or dispersed across genome.
Shutting off mutated genes by cross-linking double stranded dna
(By Nitrogen Mustard)
The supreme intellect for drug design is Ross, an Englishman, who is a Professor of Chemistry at the London University, England. Professor WCJ Ross is also the Head of Chemistry Department at the Royal Cancer Hospital, a post-graduate medical center of the London University. Ross was the first person who designed drugs for treating Cancers. He designed drugs to cross-link both strands of DNA that we inherit one strand from each parent. Cross-linking agents such as Nitrogen mustard are extremely toxic and were used as chemical weapon during the First World War (WWI). More toxic derivatives were developed during the Second World War (WWII). Using data for the toxic effect of Nitrogen Mustard on soldiers during WWI, Ross observed that Soldiers exposed to Nitrogen Mustard showed a sharp decline of White Blood Cells (WBC) that is from 5,000 cell/CC to 500 cells/CC. He immediately realized that children suffering from Childhood Leukemia have a very high WBC count that is over 90,000 cells/CC. In sick children, most of the WBCs are premature, defected, and unable to defend the body from microbial infections. Ross rationale was that cancer cells divide faster than the normal cells, by using Nitrogen Mustard to cross linking both strands of DNA, one can control and stop the abnormal WBC cell division in Leukemia patients. It was indeed found to be true. Professor Ross was the first person to synthesize many derivatives of Nitrogen Mustard. By using an analog of Nitrogen Mustard, called Chlorambucil, he was successful in treating Childhood Leukemia. In America, two Physicians named Goodman and Gilman from the Yale University were the first to use Nitrogen Mustard to treat cancer in humans. Nitrogen Mustards and its analogs are highly toxic. Ross was a Chemist, over the years, he synthesized several hundred derivatives of Nitrogen Mustard to modify toxicity of Nitrogen Mustard [6-8].
Although analogs of Nitrogen Mustard are highly toxic, they are more toxic to cancer cells and more cancer cells are destroyed than the normal cells. Toxicity is measured as the Chemotherapeutic Index (CI) which is a ratio between toxicity to Cancer cells versus the toxicity to Normal cells. Higher CI means that the drugs are more toxic to cancer cells. Most cross-linking Nitrogen Mustard have a CI of 10 that is they are ten times more toxic to cancer cells. Some of the Nitrogen Mustard analogs Ross made over the years are useful for treating cancers such as Chlorambucil for treating childhood leukemia (which brought the WBC level down to 5,000/CC). Children with Childhood Leukemia treated with Chlorambucil showed no sign of Leukemia even after 20 to 25 years later. Chlorambucil made Ross one of the leaders of the scientific world. He also made Melphalan and Myrophine for treating Pharyngeal Carcinomas[9- 13].
DNA binding aziridine group
Nitrogen Mustard neither have selectivity nor specificity. They attacked all dividing cells including normal cells. During the study of the mechanism of action of radiolabeled Nitrogen Mustard on DNA, it was discovered that the two arms of Nitrogen Mustard do not bind to the double stranded DNA simultaneously. It binds to one strand of DNA at a time. The carbonium ion of the other arm of Nitrogen mustard attacks its own Nitrogen atom forming a stable three-member aziridinium ion. (see above chart). We were unable to isolate the aziridinium ion as growing tumor which produces acid which break down aziridinium ion to produce a second carbonium ion which attacks the second strand of DNA. We were able to isolate cross-linking DNA product. This study showed that to attack a single strand of DNA, we must synthesize Aziridine in the Lab. Synthesis of Aziridine analogs will give two advantages over Nitrogen Mustard: first, instead of cross-linking both strands of DNA, Aziridine binds to one strand of DNA, reducing its toxicity of double strand Nitrogen Mustard by half. Second, it gives selectivity, the Aziridine ring opens only in the acidic medium. Once the active ingredient Aziridine was determined to attack DNA, the next question was what drug delivery method should be used to deliver Aziridine at the tumor site.
The above structures are Nitrogen Mustard (2-bischloroethyl methyl amine) and Aziridine.
DNA binding lethal groups
As a part of my doctoral thesis, I was assigned a different path. Instead of cross-linking DNA by Nitrogen Mustard, I am to design drugs to attack only one strand of DNA by making Aziridine analogues. We decided to use Aziridine moiety that would be an excellent active component to shut off a gene by binding to a single strand of DNA. To deliver Aziridine to the target site DNA, we decided to use Dinitrophenyl moiety as a delivery agent because its analog Dinitrophenol disrupt the Oxidative Phosphorylation of the ATP (Adenosine Triphosphate) which provides energy to perform all our body functions. To provide energy to our body function, the high energy phosphate bond in ATP is broken down to ADP (Adenosine Diphosphate) which is further broken down to AMP (Adenosine Mono Phosphate), the enzyme Phosphokinase put the inorganic phosphate group back on the AMP giving back the ATP. This cyclic process of Oxidative Phosphorylation is prevented by Dinitrophenol. I decided to use Dinitrophenol as drug delivery method for the active ingredient Aziridine. Dinitrophenol also serves as a dye which stains a tumor called the Walker Carcinoma 256, a solid and most aggressive tumor in Rat. The first molecule I made by attaching the C-14 radiolabeled Aziridine to the dinitrophenol dye. The Dinitrophenyl Aziridine was synthesized using Dinitrochlorobenzene with C-14 radiolabeled Aziridine in the presence of Triethyl amine which removes the Hydrochloric Acid produced during the reaction. When the compound Dinitrophenyl Aziridine was tested against the implanted experimental animal tumor, the Walker Carcinoma 256 in Rats, it showed a TI (Therapeutic Index) of ten. The TI was like most of the analogs of Nitrogen Mustard. Since this Aziridine analog was not superior to Nitrogen Mustard, it was dismissed as unimportant.
Structure Activity Relationship
Reexamination of the X-ray photographs showed that most of the radioactivity was concentrated at the injection site. Very little radioactivity was observed at the tumor site. It was obvious that we need to make derivatives of Dinitrophenyl Aziridine to move the drug from the injection site to the tumor site. Because of the lack of an effective drug delivery method, Dinitrophenyl Aziridine stays at the injection site. A very small amount of radioactivity was found on the tumor site.
Dinitrophenyl benzamide a novel drug delivery molecule for Aziridine
I immediately realized that by making water and fat-soluble analogs of Dinitrophenyl Aziridine, I should be able to move the drug from the injection site to the tumor site. To deliver 2,4-Dinitrophenylaziridine form the injection site to tumor site, I could alter the structure of 2,4-Dinitrophenylaziridine by introducing the most water-soluble group such as ethyl ester to least water-soluble group such as Cyano- group or to introduce an intermediate fat/water double Amido group.
An additional substituent in the Dinitrophenyl Aziridine could give three isomers, Ortho, Meta, and Para substituent. Here confirmational chemistry plays an important role in drug delivery. Ortho substituent always give inactive drug. Model building showed that because of the steric hinderance, Aziridine could not bind to DNA shutting off the genes. On the other hand, Meta and Para substituents offer no steric hindrance and drug could be delivered to DNA. The following chart showed that I synthesized all nine C-14 radiolabeled analogs of 2,4-Dinitrophenyl aziridines and tested them against implanted Walker Carcinoma 256 in Rats.
Derivatization of Dinitro phenyl Benzamide based on Partition Coefficient
The most water-soluble substituent
The first three compounds on top line of the above chart carry all three isomer of most water-soluble Ethyl Ester group attached to 2,4-Dinitropehny aziridine. The compound invivo is hydrolyzed ester to produce most water-soluble carboxylic group. Within 24 hours of injection, the entire radioactive compound was extracted from the Rat’s urine washed down from the cages. Since the Ortho position was not available for DNA binding, it showed no biological activity, but the third compound in which one of the Ortho position was free to bind to DNA showed some activity.
The least water-soluble substituent
On the other hand, when the least water-soluble Cyano-group was attached to all three isomers of the 2,4-Dinitrophenyl aziridine compound as shown in the second line of the above chart, most of the compound stayed at the injection site. Only the last Cyano-derivative attached to DNA showed some activity.
The moderately soluble substituent
The last line of the above chart showed that the first two Amido groups were sterically hindered and did not bind to DNA and showed no biological activity, but the last compound presents the perfect drug delivery method. The entire drug was delivered from the injection site to the tumor site. The drug 1-Aziridine, 2,4-dinitro, 5-benzamide (CB1954) showed the highest biological activity. It has a CI of seventy; it is seventy times more toxic to cancer cells, highest toxicity ever recorded against Walker Carcinoma 256 in Rats [14-16].
Nitrogen Mustards are highly toxic because they have neither specificity nor selectivity. They attack all dividing cells whether they are normal or abnormal. On the other hand, the analogs of Aziridines and Carbamates serve as prodrug and remain inactive in the basic and neutral media. They become activated only in the presence of acid producing cancer cells. Aziridine attacks DNA in acidic medium, particularly the N-7 Guanine. The dye Dinitro benzamide has great affinity for Walker Tumor. The Aziridine Dinitro benzamide (CB1954) stain the tumor. As the tumor grows, it uses Glucose as a source of energy. Glucose is broken down to Lactic Acid. It is the acid which activates the Aziridine ring. The ring opens to generate a carbonium ion which attacks the most negatively charged N-7 Guanine of DNA (as shown below) shutting off the Walker Carcinoma gene in Rat. The following conjugate structure show how CB1954 binds to a single stranded of DNA shutting off the gene.
Conjugated DNA Disrupting Protein Synthesis Pathway of Cancer Cell
For the discovery of CB1954, The University of London, honored with the Institute of Cancer Research (ICR) post-doctoral award to synthesize more analogs of CB1954. To improve drug delivery method, over the years, I made over a hundred additional analogs of Dinitro phenyl aziridine. To continue my work, I was honored with the Institute of Cancer Research Post-Doctoral Fellowship Award of the Royal Cancer Hospital of London University. To increase the toxicity of CB1954 to Walker Carcinoma, I made additional 20 analogs as a postdoctoral fellow. When I attached one more Carbonium ion generating moiety, the Carbamate moiety to the Aziridine Dinitrobenzene, the compound Aziridine Dinitro benzamide Carbamate was so toxic that its Therapeutic Index could not be measured. Although it is one of the most toxic compounds ever made, it is most useful in identifying and removing the resistant cell lines from the Tumor Bank. Further work at London University was discontinued for safety reasons. [17-18].
The Best and the Worst Dinitro phenyl Aziridine Analogs
I continued my work on the highly toxic Aziridine/Carbamate combination in America when I was offered the Fogarty International Fellowship Award to continue my work at the National Cancer Institute (NCI) of the National Institutes of Health (NIH). I brought the idea from London University of attacking one strand of DNA using not only Aziridine, but also Carbamate without using the same dye Dinitro benzamide. My greatest challenge at NCI is to translate the animal work which I did in London University to humans.
Translating Animal Work to Humans
Designing drugs to treat Glioblastoma: the human brain cancers
One day, I heard an afternoon lecture at the NIH in which the speaker described that radio labeled Methylated Quinone crosses the Blood Brain Barrier (BBB) in mice. When injected in mice, the X-ray photograph showed that the entire radioactivity was concentrated in the Mice’s brain within 24 hours. I immediately realized that Glioblastoma multiforme, the brain tumor in humans, is a solid aggressive tumor like Walker Carcinoma in Rats. I decided to use Quinone moiety as a novel drug delivery molecule to cross BBB delivering Aziridine rings to attack Glioblastomas. By introducing an additional Carbamate moiety, I could increase its toxicity several folds. I planned to use this rationale to translate animal work to human by introducing multiple Aziridine and Carbamate moieties to the Quinone to test against Glioblastomas in humans.
Glioblastoma (GBM) is a primary type of brain cancer which originates in the brain, rather than traveling to the brain from other parts of the body, such as the lungs or breasts. GBM is also called glioblastoma multi for me which is the most common type of primary brain cancer in adults. Attaching Nitrogen Mustard group to Quinone will produce highly toxic compound which will have neither specificity nor selectivity. Such a compound will attack all dividing cells whether they are normal or abnormal. On the other hand, the analogs of Aziridines and Carbamates are pro-drugs and remain inactive in the basic and neutral media. They become activated only in the presence of acid producing cancer cells.
DNA Binding Aziridines
Dr. Khan is the Discoverer of AZQ (US Patent 4,146,622), a Novel Experimental Drug Specifically Designed to shut off a Gene that causes Brain Cancer for which he receives a 17-year Royalty for his invention (License Number L-0I9-0I/0). To this date, more than 300 research papers have been published on AZQ. The award ceremony was broadcast live worldwide by the Voice of America (VOA). Dr. Khan is the first Indian to receive one of America’s highest awards in Medicine.
Exhibit #2
Exhibit #3
His Excellency, Dr. A.P.J. Abdul Kalam, The President of India Greeting. Dr. A. Hameed Khan.
Discoverer of anti-cancer AZQ, after receiving 2004, Vaidya Ratna, The Gold Medal, One Of India’s Highest Awards in Medicine. At The Rashtrapathi Bhavan (Presidential Palace), in Delhi, India, during a Reception held on April 2, 2004.
India’s National Medal of Honor
Exhibit # 4
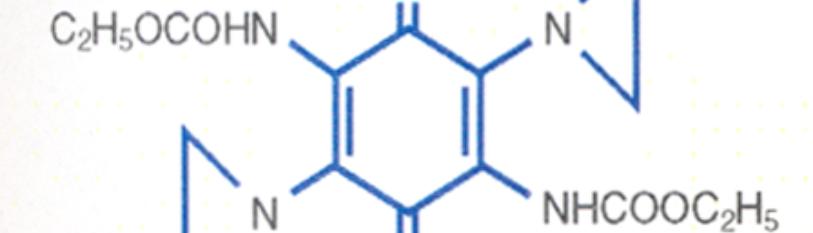
Single Strand DNA Binding Aziridine and Carbamate.
DNA Single Strand Binding Agents DNA Single Strand Binding Agents.
|
U.S. Patent 4,146,622 |
Structure of AZQ for treating Glioblastoma
Exhibit # 5
Gold Medal for Dr.Khan
Dr. A. Hameed Khan, a Scientist at the National Institutes of Health (NIH) USA, an American Scientist of Indian Origin was awarded on April 2, 2004. Vaidya Ratna; The gold Medal, one of India’s Highest Awards in Medicine for his Discovery of AZQ (US Patent 4,146,622) which is now undergoing Clinical Trials for Treating Bran Cancer.For Discovering AZQ Dr. Khan was Awarded A Gold Medal
Exhibit #6

