Proteins in the Spotlight: Unraveling the Signaling Cascade of EGFR in Cancer Development and Progression
Emad Fadhal*
Department of Mathematics & Statistics, College of Science, King Faisal University, Saudi Arabia
*Corresponding author: Emad Fadhal, Department of Mathematics & Statistics, College of Science, King Faisal University, Saudi Arabia.
Citation: Fadhal E. (2023 Proteins in the Spotlight: Unraveling the Signaling Cascade of EGFR in Cancer Development and Progression. J Can Ther Res. 3(1):1-8.
Received: August 15, 2023 | Published: August 29, 2023
Copyright© 2023 Genesis Pub by Fadhal E. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International License (CC BY 4.0). This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are properly credited.
DOI: https://doi.org/10.52793/JCTR.2023.3(1)-30
Abstract
Epidermal growth factor receptor (EGFR) signaling plays an important role in cancer by regulating cell growth, proliferation, and survival. Dysregulation of EGFR signaling leads to uncontrolled cell division and tumor progression in various cancer types. Studies on EGFR targeting have shown promise as a cancer treatment strategy.
In this study, several genes such as SHC1, SOS1, PIK3CA, EGFR, GRB2, and PIK3R1 were identified that are involved in the EGFR signaling pathway in cancer and play important roles in cancer progression and progression. It is a possible target for treatment. They are participating in various signaling pathways that regulate cell growth, survival, and metastasis. Understanding and targeting EGFR signaling has great potential to advance cancer treatment and treatment through the development of personalized targeted therapies.
Keywords
EGFR signaling pathway; Oncogenes; Suppressor gene; Protein-protein interaction networks; Cancer development
Introduction
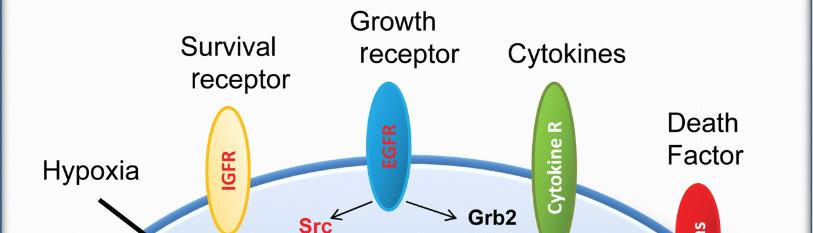
The EGFR is a crucial transmembrane receptor tyrosine kinase that is engaged in the cellular growth regulation, proliferation, and survival. Aberrant activation and signaling have been extensively linked to advancement and evolution of different cancer types. Aberrant activation of EGFR signaling pathways has been widely involved in cancer growth and progression. Upon ligand binding, EGFR undergoes autophosphorylation at specific tyrosine residues, initiating a cascade of intracellular signaling events. This leads to the activation of the subsequent signaling cascades, encompassing the ras-MAPK pathway and the PI3K-Akt pathway, which promote cell proliferation, survival, and migration. Additionally, the Janus kinase-STAT pathway can be activated by EGFR, contributing to cancer cell survival and immune evasion [1-3].
Somatic mutations in EGFR are frequent in certain cancer types, particularly non-small cell lung cancer (NSCLC) and colorectal cancer. These mutations often result in constitutive activation of EGFR, driving uncontrolled cell proliferation and oncogenic transformation. The most common EGFR mutations, such as exon 19 deletions and the L858R point mutation, have significant clinical implications as they predict responsiveness to targeted therapies such as EGFR tyrosine kinase inhibitors (TKIs). Understanding the mutational landscape of EGFR in cancer has revolutionized precision medicine, allowing for more personalized treatment strategies [4, 5].
Given the pivotal role of EGFR in cancer progression, therapeutic interventions targeting this receptor have been explored extensively. EGFR-specific TKIs, monoclonal antibodies, and combination therapies have shown promising results in clinical trials, offering improved outcomes for certain patient subgroups. Nevertheless, the heterogeneity of EGFR-driven tumors and the emergence of resistance remain substantial challenges in achieving long-term treatment success. Developing strategies to overcome these obstacles is a critical area of ongoing research [6, 7].
EGFR signaling is critically involved in the development and progression of diverse cancer types and contributes to tumor initiation, growth, and metastasis. Understanding the intricacies of EGFR signaling and its interactions with other cellular pathways is essential for devising effective therapeutic approaches that can counteract the relentless progression of cancer. Therefore, the aim of this study was to identify key proteins involved in the EGFR signaling pathway and to highlight their functions. This information provides insight into the underlying development and progression of cancer, and ultimately helps to develop more effective cancer treatments in the future.
Materials and Methods
In our previous work, we utilized a specific approach to analyze protein-protein interactions (PPIs) by treating them as a metric space and employing graph theory tools [8]. This involved considering all PPIs as a metric space and determining the topological center of each network. According to their graph-theoretical proximity to the central protein, we classified the remaining proteins into different 'zones'. For instance, zone 1 consisted of proteins one-step away from the topological center, and zone 1 included proteins two steps away.
Our results indicated that proteins interacted in a radial manner through a central node, with a high concentration of proteins near the center and a lower concentration as we move away from it. Additionally, the distribution of proteins from the center in specific hierarchies held significant biological importance.
Therefore, we concluded that formally and concisely evaluating PPIs as a metric space, with an emphasis on zones in relation to the center, can unveil crucial distinctions between PPI networks observed in normal and diseased tissues. We suggested that centrally localized proteins, specifically those implicated in sensory functions, could be promising therapeutic targets and should be subject to formal evaluation in subsequent investigations utilizing our metric space methodology. Furthermore, our continuous study has explored possible uses of this strategy, indicating that the core zones of certain human PPIs are rich in essential biomolecules and established therapeutic targets. The core zones exhibited even higher enrichment, supporting the hypothesis that they could be valuable in drug target discovery [9-12].
Based on an extension of previous analytical studies, the current study focuses on zone one, which is extremely interconnected and enriched with proteins participating in various cellular functions, such as signaling pathways, immune system responses, blood clotting processes, and disease-related pathways.
Analysis of pathways and functional enrichment
In order to assess the biological significance of distinct zones within the PPI network, we classified proteins based on their distance from the center. Subsequently, we conducted an overrepresented pathway analysis to identify particular characteristics associated with these zones. To perform this analysis, we utilized different web services, such as gene set enrichers for comparative toxicogenomics databases and gene ontology term enrichment analysis. To establish statistical significance, a threshold of 0.01 was set. Finally, we determined the ratio of proteins present in each enriched signaling pathway to determine if the zones displayed functional specialization.
An assessment of pathways involving oncogenes and tumor suppressor proteins
The protein scores underwent a thorough evaluation, particularly focusing on those observed in oncogenes and tumor suppressors. The analysis involved utilizing data from cancer genome-wide sequencing studies to pinpoint enhanced signaling pathways. A key aspect of the investigation was examining interactions that exhibited high scores. The results unveiled a notable trend: these interactions predominantly involved genes causally linked to cancer [13].
Crucial proteins involved in cellular processes, signaling, growth, cell cycle regulation, and with potential as therapeutic targets.
For assessing zone 1 of the human functional protein interaction network, we employed a collection of vital human proteins identified based on the corresponding gene knock-out effects observed in mice [14,15].
Ethics approval of research
We would like to clarify that the research article in question does not involve human participants or patient material. The study primarily relies on data from established databases and does not directly involve any interaction, intervention, or collection of information from individuals. It is purely a research article based on existing data sources, with no direct involvement of human subjects.
Results
Our study focused on 374 proteins in zone 1 and examined their involvement in various signaling pathways related to EGFR in cancer using the KEGG pathway database [16]. Activation and regulation of these signaling pathways are critical for cancer development and progression. However, successful targeting of cancer cell signaling pathways is often hampered by alterations and dysregulation within these pathways that promote cancer cell proliferation and survival. Disruption of these signaling pathways can impair their ability to support cancer growth [17-21].
Among the proteins analyzed (SHC1, SOS1, PIK3CA, EGFR, GRB2, PIK3R1), signaling proteins accounted for the largest proportion, accounting for 100% of the total, signaling proteins being the most important over other functions was emphasized. Essential proteins follow at 83.3%. The remaining functions are expressed in the following ratios: MAPK cascade (33.3%), negative signal (33.3%), positive signal (16.6%), and cell cycle (16.6%).
Proteins play important roles in cancer networks, functioning as both cancer-promoting oncogenes and cancer-preventing tumor suppressor genes. Their interactions strongly influence cancer cell behavior [22, 23]. Remarkably, 33.3% of the proteins were identified as oncogenes, including PIK3CA and EGFR. This highlights the importance of specific zones with high protein concentrations, some of which may be potential drug targets. Another function of the protein in this context is tumor suppression (PIK3R1) (16.6%). Our results identify several key proteins in the EGFR signaling pathway that play critical roles in cancer development and progression. Here we introduce them and their importance in cancer. SHC1 plays an important role in carcinogenesis as it functions as an adapter protein, connecting activated receptors to downstream signaling pathways and promoting cell proliferation and survival [24]. The guanine nucleotide exchange factor SOS1 is essential for cancer progression because it activates the small GTPase Ras, which initiates a cascade of signaling events that lead to uncontrolled cell proliferation and tumorigenesis [25]. PIK3CA, the catalytic subunit of phosphoinositide-3-kinase (PI3K), is frequently mutated in various types of cancers and promotes activation of the PI3K-Akt signaling pathway, thus enhancing cancer cell survival; It contributes to proliferation and metastasis [26]. EGFR, a cell surface receptor tyrosine kinase, is a well-known oncogenic driver in several types of cancer because its defective activation leads to increased resistance to cell proliferation, angiogenesis, and apoptosis [27]. The adapter protein GRB2 is important in cancer progression as it mediates downstream signaling of multiple growth factor receptors and transmits oncogenic signals that support cancer cell survival and proliferation [28]. PIK3R1, also known as p85α, is the regulatory subunit of PI3K. Dysregulation of PIK3R1 in cancer contributes to sustained stimulation of the PI3K-Akt signaling cascade, promoting malignant tumor growth, invasion, and metastatic spread [29].
Proteins within the EGFR signaling pathway exert significant influence over crucial cellular and disease-related pathways
To assess the significance of pathway enrichment among proteins in the EGFR signaling pathway compared to the proteins present in specific KEGG pathways, we conducted a mapping process, aligning proteins from the EGFR signaling pathway to those within the respective KEGG pathways. The entire set of proteins present in the EGFR signaling pathway participates in KEGG pathways such as 'pathways in cancer,' 'breast cancer,' 'Endocrine resistance,' and 'MicroRNAs in cancer,' as illustrated in (Figures 1-4).
Figure 1: KEGG pathway in cancer.
Figure 2: KEGG breast cancer pathway.
Figure 3: KEGG Endocrine resistance pathway.
Figure 4: KEGG MicroRNAs in cancer pathway.
Discussion
Cancer is an intricate condition marked by dysregulated signaling pathways, leading to uncontrolled cell growth and proliferation. The EGFR signaling pathway is one of the principal pathways engaged in cancer development and progression. Understanding the proteins and their roles within this signaling pathway is critical for identifying potential drug targets and developing effective cancer therapeutics. In this study, 374 proteins in zone 1 were analyzed and their involvement in various signaling pathways related to EGFR in cancer was examined using the KEGG signaling pathway database.
Signaling proteins constituted the largest proportion (100%) of the analyzed proteins, highlighting their paramount importance over other functions. Signaling pathways are crucial in cancer as they regulate diverse cellular activities, like, cell growth, persistence, and apoptosis. Dysregulation of these pathways can promote cancer cell growth, and disrupting them presents a promising approach for cancer treatment.
Proteins play critical roles in cancer networks, functioning as both cancer-promoting oncogenes and cancer-preventing tumor suppressor genes. The interactions between these proteins strongly influence cancer cell behavior. Notably, 33.3% of the analyzed proteins were identified as oncogenes, further emphasizing the significance of specific zones with high concentrations of proteins that could serve as potential drug targets. Additionally, 16.6% of the proteins in the study were associated with tumor suppression, suggesting their potential role in inhibiting cancer progression.
The discussed genes, SHC1, SOS1, PIK3CA, EGFR, GRB2, and PIK3R1, play pivotal roles in the advancement and progression of cancer, making them potential targets for cancer treatment. They are involved in various signaling pathways that regulate cell growth, survival, and metastasis. Inhibiting these genes or their downstream signaling pathways could lead to novel therapeutic strategies for different types of cancer [30-34].
Conclusion
This study highlights that the EGFR signaling pathway in cancer plays an important role in regulating cell growth, proliferation, and survival. Dysregulation of EGFR signaling contributes to uncontrolled cell division and tumor progression in various cancer types. The identification of key genes involved in this signaling pathway, including SHC1, SOS1, PIK3CA, EGFR, GRB2 and PIK3R1, suggests their potential as therapeutic targets for cancer therapy. By understanding and targeting EGFR signaling, we can develop personalized targeted therapies to advance cancer treatment. The results of this study move us closer to more strategies that are effective in the fight against cancer.
Acknowledgments
We would like to thank Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia for support. The Deanship of Scientific Research, Vice Presidency supported this work for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia.
Conflict of interest
The author declare there is no conflict of interest.
References
- Perrone F, De Laurentiis M, De Placido S, Orditura M, Cinieri S, et al. (2019) Adjuvant zoledronic acid and letrozole plus ovarian function suppression in premenopausal breast cancer: HOBOE phase 3 randomised trial. Eur J Cancer. 118:178-86.
- Falcone F, Normanno N, Losito NS, Scognamiglio G, Abate RE, et al. (2019) Application of the Proactive Molecular Risk Classifier for Endometrial Cancer (ProMisE) to patients conservatively treated: Outcomes from an institutional series. Eur J Obstet Reprod Biol. 240:220-25.
- Yarden Y, Pines G. (2012) The ERBB network: at last, cancer therapy meets systems biology. Nat Ren Cancer. 12(8):553-63.
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, et al. (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N EnglJ Med. 350(21):2129-39.
- Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, et al. (2017) Alectinib versus crizotinib in untreated ALK-positive non–small-cell lung cancer. N Engl J Med. 377(9):829-38.
- Singh R, Kumar S, Liu FZ, Shuang C, Zhang B, et al. (2020) Etched multicore fiber sensor using copper oxide and gold nanoparticles decorated graphene oxide structure for cancer cells detection. Biosensors and Bioelectronics. 2020 Nov 15;168(6):112-557.
- SequistLV Wm, Turke BA, Gettiniger S, Lynch JT, Temel J, et al. (2011) Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 3(75):75ra26.
- Fadhal E, Gamieldien J, Mwambene EC. (2014) Protein interaction networks as metric spaces: a novel perspective on distribution of hubs. BMC Syst Biol. 8(1):1-1.
- Fadhal E, Mwambene EC, Gamieldien J. (2014) Modelling human protein interaction networks as metric spaces has potential in disease research and drug target discovery. BMC Syst Biol. 8(1):1-2.
- Fadhal E. (2023) Unraveling the significance of signal transduction pathways: Key players in cancer development and progression. J Can Ther Res. 3(1):1-9.
- Fadhal E. (2023) Exploring the significance of zone 1 in human protein interaction networks: Identifying potential therapeutic targets for cancer treatment. Oncology and Radiotherapy. 17 (8) 249-53.
- Fadhal E, Gamieldien J, Mwambene EC. (2015) Self-similarity of human protein interaction networks: a novel strategy of distinguishing proteins. Scientific Reports. 5(1):7628
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz Jr LA, et al. (2013) Cancer genome landscapes. science. 339(6127):1546-58.
- Wu G, Feng X, Stein L. (2010) A human functional protein interaction network and its application to cancer data analysis. Genome biol. 11(5):1-23
- Bult CJ, Eppig JT, Blake JA, Kadin JA, Richardson JE, et al. (2012) Mouse Genome Database Group. The mouse genome database: genotypes, phenotypes, and models of human disease. Nucleic acids research. 41(D1):885-91.
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, et al. (2019) The comparative toxicogenomics database: update 2019. Nucleic acids research. 47(D1):D948-54.
- Yang Y, Li X, Wang T, Guo Q, Xi T, et al. (2020) Emerging agents that target signaling pathways in cancer stem cells. Journal of hematology & oncology. 13:1-8.
- Nouri Z, Fakhri S, Nouri K, Wallace CE, Farzaei MH, et al. (2020) Bishayee A. Targeting multiple signaling pathways in cancer: The rutin therapeutic approach. Cancers. 12(8):2276.
- Clara JA, Monge C, Yang Y, Takebe N. (2020) Targeting signalling pathways and the immune microenvironment of cancer stem cells—A clinical update. Nat rev clin oncol. 17(4):204-32.
- He Y, Xu W, Xiao YT, Huang H, Gu D, et al. (2022) Targeting signaling pathways in prostate cancer: Mechanisms and clinical trials. Signal transduct target ther. 7(1):198.
- Levine AJ, Puzio-Kuter AM. (2010) The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 330(6009):1340-4.
- Aunoble BÉ, Sanches R, Didier E, Bignon YJ. (2000) Major oncogenes and tumor suppressor genes involved in epithelial ovarian cancer. Int j oncol. 16(3):567-643.
- Golkaram M, Kuo F, Gupta S, Carlo MI, Salmans ML, et al. (2022) Spatiotemporal evolution of the clear cell renal cell carcinoma microenvironment links intra-tumoral heterogeneity to immune escape. Genome Medicine. 14(1):143
- Hirano F, Kaneko K, Tamura H, Dong H, Wang S, et al. (2005) Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer res. 65(3):1089-96.
- Schubbert S, Shannon K, Bollag G. (2007) Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 7(4):295-308.
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, et al. (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science. 304(5670):554-.
- Mendelsohn J, Baselga J. (2003) Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J clin oncol. 21(14):2787-99.
- Parent JM, Lowenstein DH. (2002) Seizure-induced neurogenesis: are more new neurons good for an adult brain. Prog brain res. 135:121-31.
- Li W, Ge Z, Liu C, Liu Z, Björkholm M, et al. (2008) CIP2A is overexpressed in gastric cancer and its depletion leads to impaired clonogenicity, senescence, or differentiation of tumor cells. Clin cancer Res. 14(12):3722-8.
- Shahrokni A, Tin A, Alexander K, Sarraf S, Afonso A, et al. (2019) Development and evaluation of a new frailty index for older surgical patients with cancer. JAMA Netw Open. 2(5):e193545-.
- Lv Y, Zhao SG, Lu G, Leung CK, Xiong ZQ, et al. (2017) Identification of reference genes for qRT-PCR in granulosa cells of healthy women and polycystic ovarian syndrome patients. Scientific reports. 7(1):6961.
- Selven H. (2020) Assessing new prognostic biomarkers in resected colon cancer patients. Cancers.12(2):319
- Yarden Y, Pines G. (2012) The ERBB network: at last, cancer therapy meets systems biology. Nature Reviews Cancer. 12(8):553-63.
- Hao wang. (2020) Targeting EphA2 in Hepatocellular Carcinoma: Uncovering Its Therapeutic Potential and Beyond." PhD diss., Loyola University Chicago.

